Mini-review: Macrophage Polarization
Macrophages were originally identified by Ilya Metchnikoff (Nathan CF 2008) more than 100 years ago, and his description of phagocytosis won him the Nobel Prize for Medicine in 1908. Macrophages are a heterogeneous population of innate myeloid cells involved in health and disease. They are the most functionally diverse (plastic) cells of the hematopoietic system, found in all tissues, and their main function is to respond to pathogens and modulate the adaptive immune response through antigen processing and presentation. Further functions of macrophages center on the induction and resolution of inflammation, as well as tissue repair (Mosser DM and Edwards JP 2008). Macrophages have different functions and transcriptional profiles, but all are required for maintaining homeostasis. This involves phagocytosis of debris and pathogens, dead cell clearance, and matrix turnover. Here we provide an overview of macrophage polarization, focusing on the characterization and function of the various macrophage subsets.
History of the terminology of macrophage polarization
In the 1990s it was discovered that the cytokine interleukin (IL)-4 induced different effects on macrophage gene expression compared to that of interferon (IFN)-gamma and lipopolysaccharide (LPS). In contrast to the classical activation of macrophages by IFN-gamma (Nathan CF et al. 1983), the macrophage gene expression induced by IL-4 was described as “alternative activation” (Stein M et al. 1992, Martinez FO and Gordon S 2014).
A few years later, in 2000, Mills et al. proposed a new classification of macrophages as either M1 or M2. This terminology originated from an observation of differential macrophage arginine metabolism in various mouse strains with T helper type 1 (Th1) (C57BL/6 mice) and T helper type 2 (Th2) (Balb/c mice) backgrounds. Th1 mice with T cells producing mostly IFN-gamma demonstrated macrophage activation in which nitric oxide (NO) was generated from arginine, versus ornithine production from Th2 mice with T cells producing IL-4 and tumor growth factor (TGF)-beta 1 (Mills CD et al. 2000). This finding led to a consensus within the scientific community that M1 (classically activated) macrophages exhibit inflammatory functions, whereas M2 (alternatively activated) macrophages exhibit anti-inflammatory functions. In 2004, Mantovani et al. further divided M2 macrophages into M2a, M2b, M2c and M2d subtypes based on the applied stimuli and the induced transcriptional changes (Mantovani A et al. 2004, Rőszer T 2015).
The M1/M2 classification of macrophages is now considered an oversimplified approach that does not adequately describe the spectrum of macrophage populations. For instance, the identification of tumor associated macrophages (TAMs), which do not fit nicely into the criteria for M1 or M2 macrophages complicates this system (Qian BZ and Pollard JW 2010). In addition, macrophages expressing T cell receptors (TCR) and CD169 have also been identified.
Phenotype of M1 and M2 macrophage subsets and their functions
Macrophages derived from monocyte precursors undergo specific differentiation depending on the local tissue environment. They respond to environmental cues within tissues such as damaged cells, activated lymphocytes, or microbial products, to differentiate into distinct functional phenotypes. The M1 macrophage phenotype is characterized by the production of high levels of pro-inflammatory cytokines, an ability to mediate resistance to pathogens, strong microbicidal properties, high production of reactive nitrogen and oxygen intermediates, and promotion of Th1 responses. In contrast, M2 macrophages are characterized by their involvement in parasite control, tissue remodeling, immune regulation, tumor promotion and efficient phagocytic activity.
LPS, IFN-gamma and granulocyte-macrophage colony stimulating factor (GM-CSF) polarize macrophages towards the M1 phenotype, which induces secretion of large amounts of cytokines such as IL-1-beta, tumor necrosis factor (TNF), IL-12, IL-18 and IL-23. This helps to drive antigen specific Th1 and Th17 cell inflammatory responses. Phenotypically, M1 macrophages express high levels of major histocompatibility complex class II (MHC II), the CD68 marker, and co-stimulatory molecules CD80 and CD86 (see Table 1). M1 macrophages have also been shown to up-regulate the expression of the intracellular protein suppressor of cytokine signaling 3 (SOCS3), as well as activate inducible nitric oxide synthase (NOS2 or iNOS) to produce NO from L-arginine (MacMicking J et al. 1997, Arnold CE et al. 2014) (Table 1). In disease contexts, M1 macrophages are implicated in initiating and sustaining inflammation, and can therefore be detrimental to health.
In contrast, M2 macrophage activation is induced by fungal cells, immune complexes, helminth infections, complement components, apoptotic cells, macrophage colony stimulating factor (MCSF), IL-4, IL-13, IL-10 and TGF-beta. This activation leads to the secretion of high amounts of IL-10 and low levels of IL-12. Phenotypically M2 macrophages have been characterized as IL-12lowIL-10highIL-1decoyRhighIL-1RAhigh. They are also defined as IL-2low, IL-23low, IL-1betalow and caspase-1low. In addition, they express high levels of scavenger mannose and galactose E-type and C-type receptors (Table 1), and repurpose arginine metabolism to express ornithine and polyamine, which promotes growth.
It is now appreciated that the M2 terminology encompasses a functionally diverse group of macrophages rather than a unique activation state. Accordingly, M2 macrophages can be further divided into subsets (Table 1), specifically M2a, M2b, M2c and M2d based on their distinct gene expression profiles (Mantovani A et al. 2004, Rőszer T 2015). The M2a subtype is elicited by IL-4, IL-13 or fungal and helminth infections. M2b is elicited by IL-1 receptor ligands, immune complexes and LPS whereas M2c is elicited by IL-10, TGF-beta and glucocorticoids. The fourth type, M2d, is elicited by IL-6 and adenosine. M2d macrophages have phenotypic and functional attributes similar to ovarian TAMs but are distinct from M2a-c (Duluc D et al. 2007).
M1 and M2 macrophages have distinct chemokine and chemokine receptor profiles, with M1 secreting the Th1 cell attracting chemokines CXCL9 and CXCL10 and M2 secreting CCL17, CCL22 and CCL24. It has recently been demonstrated that in vitro, macrophages are capable of complete repolarization from M2 to M1, and can reverse their polarization depending on the chemokine environment (Davis MJ et al. 2013). The change in polarization is rapid and involves rewiring of signaling networks at both the transcriptional and translational levels.
Table 1. Classically activated (M1) and alternatively activated (M2) subset phenotypes
| M1 | M2a | M2b | M2c | M2d | |
|---|---|---|---|---|---|
| Stimulation/Activation |
IFN-gamma LPS GM-CSF |
IL-4 IL-13 Fungal and Helminth infection |
ICs IL-1R |
IL-10 TGF-beta GCs |
IL-6 LIF Adenosine |
| Marker Expression |
CD86 CD80 CD68 MHC II IL-1R TLR2 TLR4 iNOS SOCS3 |
CD163 MHC II SR MMR/CD206 CD200R TGM2 DecoyR IL-1R II Mouse only: Ym1/2 Fizz1 Arg-1 |
CD86 MHC II |
CD163 TLR1 TLR8 |
VEGF |
| Cytokine secretion |
TNF IL-1 beta IL-6 IL-12 IL-23 |
IL-10 TGF-beta IL-1ra |
IL-1 IL-6 IL-10 TNF-alpha |
IL-10 TGF-beta |
IL-10 IL-12 TNF-alpha TGF-beta |
| Chemokine secretion |
CCL10 CCL11 CCL5 CCL8 CCL9 CCL2 CCL3 CCL4 |
CCL17 CCL22 CCL24 |
CCL1 | CCR2 |
CCL5 CXCL10 CXCL16 |
Adapted from Rőszer T 2015 and Duluc D et al. 2007. Arg-1, arginase-1; FIZZ1, resistin-like molecule-alpha (Relm-alpha); GCs, glucocorticoids; ICs, immune complexes; IL1-ra, IL-1 receptor antagonist; LIF, leukocyte inhibitory factor; TGM2, transglutaminase 2; TGF-beta, transforming growth factor-beta; TNF-alpha, tumor necrosis factor alpha; TLR, Toll-like receptor; MMR (CD206), macrophage mannose receptor; iNOS, inducible nitric oxide synthase; SR, scavenger receptor; SOCS3, suppressor of cytokine signaling 3; VEGF, vascular endothelial growth factor; Ym1 (also known as chitinase-3-like protein-3 (Chi3l3)).
Signaling molecules involved in M1/M2 polarization
A network of transcription factors and post-transcriptional regulators are involved in M1/M2 polarization (Sica A and Mantovani A 2012) (Figure 1). Interferon regulatory factor (IRF), signal transducers and activators of transcription (STAT) and suppressor of cytokine signaling (SOCS) proteins all play a role in skewing macrophage function towards either the M1 or M2 phenotype. The IRF/STAT pathways, activated by IFNs and toll-like receptor (TLR) signaling, polarize macrophages to the M1 activation state via STAT1. On the other hand, IL-4 and IL-13 skew macrophages toward the M2 activation state via STAT 6 (Sica A and Bronte V 2007).
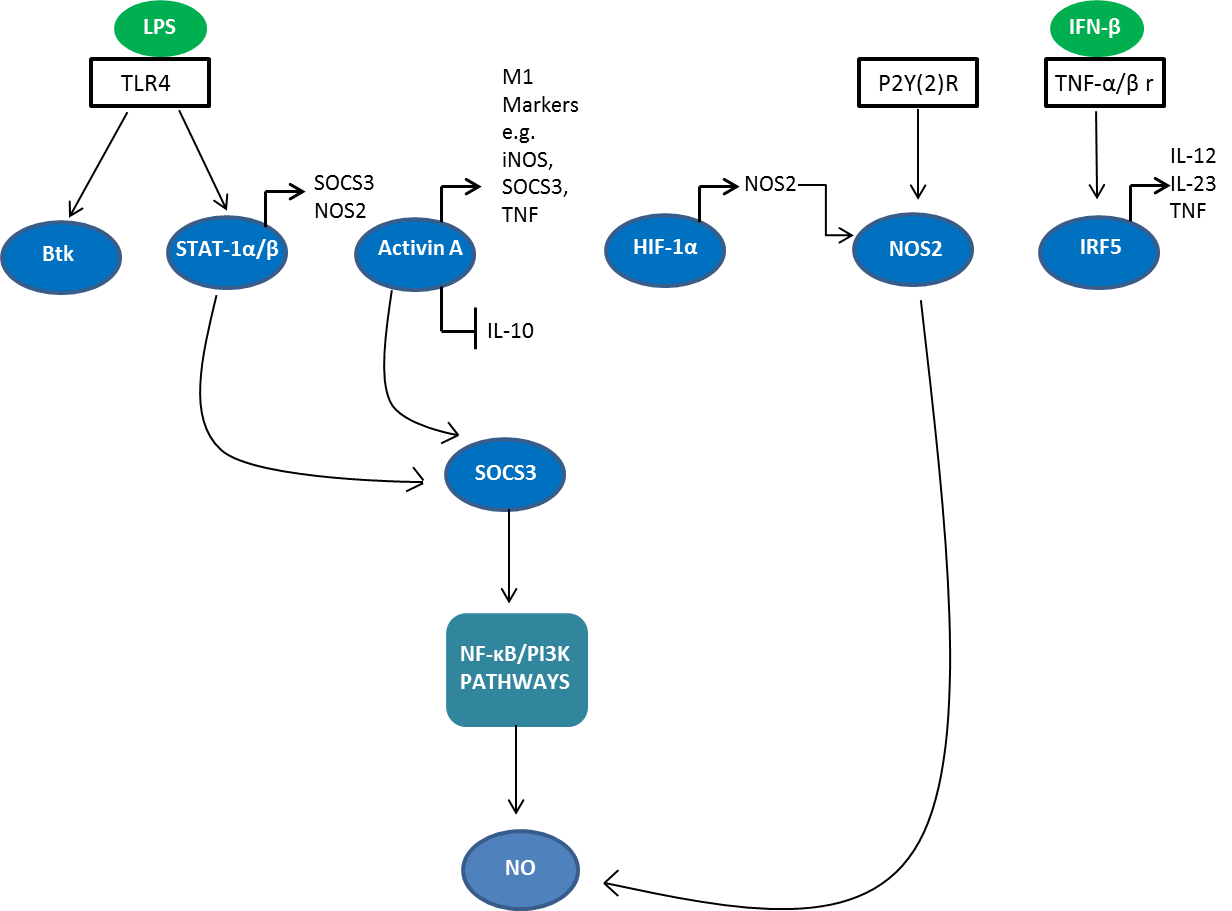
Fig. 1. Signaling molecules involved in M1 polarization. STAT, signal transducers and activators of transcription; IRF, interferon regulatory factor; SOCS, suppressor of cytokine signaling 3; Btk, Bruton’s tyrosine kinase; HIF-1, hypoxia inducible factor 1; TNF-alpha, tumor necrosis factor; iNOS, inducible nitric oxide synthase; NOS2, nitric oxide synthase 2; NF-κB, nuclear factor-kappa B; NO, nitric oxide; PI3K, phosphatidyl inositol 3 kinase; TLR4, Toll-like receptor 4; LPS, lipopolysaccharide.
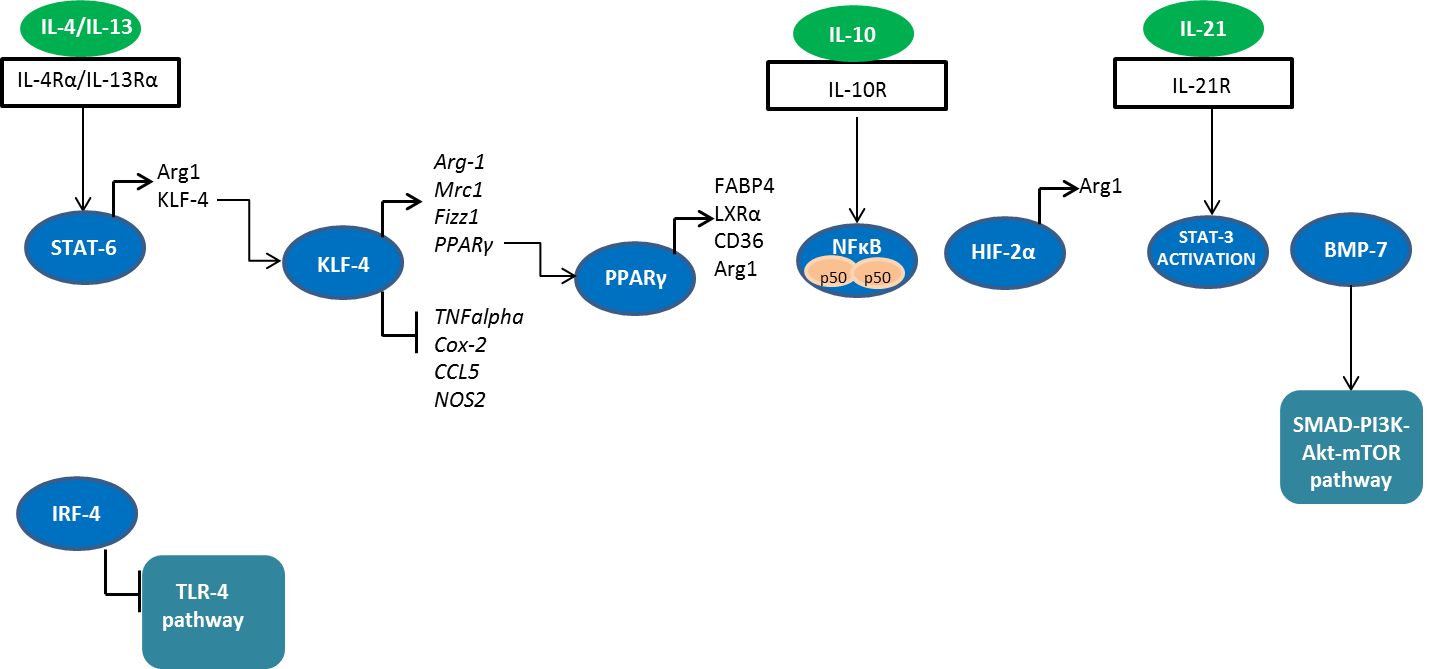
Fig. 2. Signaling molecules involved in M2 polarization. STAT, signal transducers and activators of transcription; IRF, interferon regulatory factor; HIF-2, hypoxia inducible factor 2; iNOS, inducible nitric oxide synthase; NF-κB, nuclear factor-kappa B; PI3K, phosphatidyl inositol 3 kinase; TLR4, Toll-like receptor 4; Arg-1; arginase 1; KLF-4; Krüppel-like factor 4; FIZZ1, resistin-like molecule-alpha (Relm-alpha); BMP-7, bone morphogenetic protein 7; PPARγ, peroxisome proliferator-activated receptor γ; FABP4, fatty acid binding protein 4; LXRα; liver X receptor alpha.
Table 2. Signaling molecules and genes involved in each macrophage polarization state
| M1 | M2 | |
|---|---|---|
| Signaling Molecules |
STAT1alpha/beta IRF5 Btk P2Y(2)R SOCS3 Activin A HIF1-α |
STAT6 IRF4 KLF-4 NF-κB p50 homodimers PPARγ HIF-2α IL-21 BMP-7 FABP4 LXRα |
| Genes | TNF alpha, Cox-2, CCL5, NOS2 | Arg-1, Mrc-1, Fizz1, PPARγ |
Adapted from Sica A and Mantovani A 2012 and Chávez-Galán L et al. 2015. Arg-1, arginase-1; FIZZ1, resistin-like molecule-alpha (Relm-alpha); STAT, signal transducers and activators of transcription; IRF, interferon regulatory factor; SOCS, suppressor of cytokine signaling 3; Btk, Bruton’s tyrosine kinase; HIF-1, hypoxia inducible factor 1; KLF-4, Krüppel-like factor 4; TNF-alpha, tumor necrosis factor-alpha; BMP-7, bone morphogenetic protein 7; P2Y(2)R, P2Y purinoceptor 2; PPARγ, peroxisome proliferator-activated receptor γ; NF-κB, nuclear factor-kappa B; FABP4, fatty acid binding protein 4; LXRα; liver X receptor alpha.
Mouse and human macrophages- similarities and differences
Some of the markers used to define M1 and M2 polarization vary between mouse and human macrophages (Martinez FO et al. 2013). For instance, there are no human homologs of the mouse M2 markers Ym1, Fizz1, or arginase-1. However, some molecules such as the multifunctional enzyme transglutaminase 2 (TGM2) serves as conserved markers for both human and mouse M2 macrophages and monocytes (Martinez FO et al. 2013). Identification of human M2 macrophages can now easily be performed by immunohistochemistry double staining techniques using a combination of TGM2, mannose receptor C type 1 (MRC1/CD206), and CD68 antibodies.
The following table highlights some key markers for distinguishing human and mouse M1 and M2 macrophages.
Table 3. Human and mouse M1 and M2 phenotype markers
| M1 | M2 | |
|---|---|---|
| Human | CD64, IDO, SOCS1, CXCL10 | MRC1, TGM2, CD23, CCL22 |
| Mouse | CXCL9, CXCL10, CXCL11, NOS2 | Mrc1, tgm2, Fizz1, Ym1/2, Arg1 |
Adapted from Martinez FO et al. 2013 and Martinez FO and Gordon S 2014. Arg-1, arginase-1; FIZZ1, resistin-like molecule-alpha (Relm-alpha);CCL22, chemokine (C-C motif) ligand 22; MRC1, mannose receptor C Type 1; NOS2, nitric oxide synthase 2; SOCS1, suppressor of cytokine signaling 1; TGM2, transglutaminase 2; CXCL, chemokine (C-X-C motif) ligand; IDO, indoleamine 2,3-dioxygenase.
Characterization of tumor associated macrophages
Macrophages are one of the major populations of infiltrating leukocytes associated with solid tumors (Gordon S and Taylor PR 2005). They can be recruited to the tumor site from surrounding tissues by the tumor itself through secretion of chemotactic molecules. In addition, monocytes circulating in the blood stream can infiltrate into the tumor microenvironment and mature into TAMs (Kitamura T et al. 2015). Studies in mice have suggested that the tumor infiltrating monocyte pool is primarily Ly-6C+CX3CR1low, and TAM monocyte precursors are exclusively Ly-6Chigh cells (Movahedi K et al. 2010).
The term TAMs actually describes various macrophage subsets that vary depending on the cytokine balance within the tumor microenvironment They have been primarily described as having an M2-like phenotype (Sica A et al. 2006) but studies have shown that they can express both M1 and M2 polarization hallmarks (Mantovani A et al. 2006, Allavena P et al. 2008). Notably, switching TAMs to a predominantly M1 phenotype has been proposed as a key anti-cancer immunotherapeutic treatment strategy (Mills CD et al. 2016).
TAMs are known to promote tumor progression and are associated with poor prognosis (Komohara Y et al. 2014). They induce angiogenesis, lymphogenesis, stroma remodeling and immune suppression. They also play a key role in promoting tumor invasion and metastasis (Komohara Y et al. 2015) through secretion of the enzymes plasmin, uPA, matrix metalloproteinases (MMPs) and cathepsin B (Gocheva V et al. 2010, Wang R et al. 2011).
TAMs exhibit a distinct transcriptional profile from M1 and M2 macrophages (Biswas SK et al. 2006) and are phenotypically characterized as CCL2hiCCL5hiIL-10hi. They also express MGL-1, Dectin-1, CD68, CD206, VEGF-A, NOS2, CD81, MHC II and scavenger receptor A (Chávez-Galán L et al. 2015, Rőszer T 2015). In addition, they exhibit enhanced IRF-3/STAT-1 activation and defective NF-κB signaling (Biswas SK et al. 2006).
The table below shows a selection of human cancers and the TAM markers used for diagnosis.
Table 4. Markers of tumor associated macrophages in human cancers
| Organ | Cancer type | Markers | Method | References |
|---|---|---|---|---|
| Lymph node | Hodgkin’s lymphoma |
TMA IHC |
Harris JA et al. 2012 | |
| Lung | Non-small cell lung cancer (NSCLC) |
iNOS HLA-DR |
DIHC | Ohri CM et al. 2011 |
| Breast | Breast carcinoma | MAC387 | IHC | Medrek C et al. 2012 |
| Uterus | Endometrial carcinoma | IHC | Espinosa I et al. 2012 | |
| Peripheral lymphoma | Angioimmunoblastic T cell lymphoma | DIHC | Komohara ND et al. 2010 | |
| Ovarian and peritoneum | Ovarian carcinoma |
HAM56 B7-H4 |
DIHC FIHC FC |
Kryczek I et al. 2006 |
| Colon | Colon adenocarcinoma | CCL2 | IHC |
Forssell J et al. 2007 Hu H et al. 2009 |
Adapted from Heusinkveld M and van der Burg SH 2011. FC, flow cytometry; DIHC, double immunohistochemistry; FIHC, fluorescent immunohistochemistry staining; IHC, immunohistochemistry; TMA, tissue microarray.
T cell receptor+ and CD169+ macrophages
In contrast to the macrophage subsets described above, there is limited data regarding the characteristics of TCR+ and CD169+ macrophages. However, their unique traits have fascinated immunologists and further research into their roles in regulating immune function is currently underway.
As the name suggests, TCR expression was originally thought to be exclusive to T cells, however recent studies indicate that other leukocyte subsets such as neutrophils also express the receptor (Puellmann K et al. 2006). Moreover, several studies have reported the presence of both human and mouse TCR+ macrophages (Chávez-Galán L et al. 2015). These macrophages express the TCR co-receptor CD3 as well as TCRαß and γδ subtypes (Beham AW et al. 2011, Chávez-Galán L et al. 2015). TNF has been reported to be a key regulator of TCRαß expression in macrophages (Beham AW et al. 2011), and cholesterol import/export was shown to be an important modulator of expression of the TCRαß repertoire (Fuchs T et al. 2015). TCRγδ macrophages have been implicated in host defense against bacterial challenge (Fuchs T et al. 2013).
Both subsets of TCR+ macrophages express molecules shown to be necessary for T cell signaling such as ZAP70, LAT, Fyn and Lck. Furthermore, they demonstrate high phagocytic capacity and secrete the chemokine CCL2. They have also been implicated in inflammatory and infectious diseases (Chávez-Galán L et al. 2015).
CD169+ macrophages represent the other subset of unconventional macrophages. CD169, also known as sialoadhesin or sialic acid binding immunoglobulin-like lectin (Siglec) 1, is the founding member of the Siglec superfamily of proteins. CD169+ macrophages are primarily located in secondary lymphoid organs but redistribute upon immune activation. It is reported that anti-CD169 antibodies label three macrophage populations in mouse secondary lymphoid organs: marginal zone metallophilic (MZM) macrophages in the spleen, and subcapsular sinus (SS) and medullary (Med) macrophages in lymph nodes (Crocker PR and Gordon S 1989, Oetke C et al. 2006). The signaling pathways involved in the activation of this macrophage subset are not fully understood but some aspects of their biological functions have been elucidated. For instance, they do not mediate phagocytosis but are mainly involved in immune regulation rather than steady state homeostasis. Recently, CD169+ macrophages were shown to be capable of antigen presentation to B cells and activation of CD8+ T cells (Martinez-Pomares L and Gordon S 2012). Other molecules that characterize these macrophages include CD11b, MHC II, CD68, CD11c and F4/80, although F4/80 is not expressed on all CD169+ macrophages (Chávez-Galán L et al. 2015).
The current nomenclature for macrophages is complex and the identification of CD169+ and TCR+ macrophages certainly adds to the complexity. Further research is needed to fully understand the exclusive role of these macrophages in disease. The table below summarizes the markers currently used to identify these unique macrophage subsets.
Table 5. Additional molecular markers for characterizing TCR+ and CD169+ macrophages
| TCR+ | CD169+ |
|---|---|
|
LAT Lck |
F4/80 |
Adapted from Chàvez-Galàn L et al. 2015. ZAP70, zeta- chain (TCR) associated protein kinase 70 kDa; LAT, linker of activated T cells, MHC II, major histocompatibility complex class II.
References:
- Allavena P et al. (2008). The yin-yang of tumor-associated macrophages in neoplastic progression and immune surveillance. Immunol Rev 222, 155-161.
- Arnold CE et al. (2014). A critical role for suppressor of cytokine signaling 3 in promoting M1 macrophage activation and function in vitro and in vivo. Immunology 141, 96-110.
- Beham AW et al. (2011). A TNF-regulated recombinatorial macrophage immune receptor implicated in granuloma formation in tuberculosis. Plos Pathog 7, e1002375.
- Biswas SK et al. (2006). A distinct and unique transcriptional program expressed by tumor-associated macrophages (defective NF-kappaB and enhanced IRF-3/STAT1 activation). Blood 107, 843-861.
- Bouhlel MA et al. (2007). PPARgamma activation primes human monocytes into alternative M2 macrophages with anti-inflammatory properties. Cell Metab 6, 137-143.
- Chávez-Galán L et al. (2015). Much more than M1 and M2 macrophages, there are also CD169+ and TCR+ macrophages. Front Immunol 6, 253.
- Crocker PR and Gordon S (1989). Mouse macrophage hemagglutinin (sheep erythrocyte receptor) with specificity for sialylated glycoconjugates characterized by a monoclonal antibody. J Exp Med 169, 1333-1346.
- Davis MJ et al. (2013). Macrophage M1/M2 polarization dynamically adapts to change in cytokine microenvironments in Cryotococcus neoformans infection. mBio 4, 1-10.
- Duluc D et al. (2007). Tumor-associated leukemia inhibitory factor and IL-6 skew monocyte differentiation into tumor-associated macrophage-like cells. Blood 110, 4319-4330.
- Espinosa I et al. (2010). Myometrial invasion and lymph node metastasis in endometrioid carcinomas: tumor-associated macrophages, microvessel density, and HIF1A have a crucial role. Am J Surg Pathol 34, 1708-1714.
- Eun SY et al. (2014). LPS potentiates nucleotide-induced inflammatory gene expression in macrophages via the upregulation of P2Y2 receptor. Int Immunopharmacol 18, 270-276.
- Forssell J et al. (2007). High macrophage infiltration along the tumor front correlates with improved survival in colon cancer. Clin Cancer Res 13, 1472-1479.
- Fuchs T et al. (2013). A second combinatorial immune receptor in monocytes/macrophages is based on the TCRγδ. Immunobiology 218, 960-968.
- Fuchs T et al. (2015). The macrophage –TCRαß is a cholesterol-responsive combinatorial immune receptor and implicated in atherosclerosis. Biochem Biophys Res Commun 456, 59-65.
- Gocheva V et al. (2010). IL-4 induces cathepsin protease activity in tumor-associated macrophages to promote cancer growth and invasion. Genes Dev 24, 241-255.
- Gordon S and Taylor PR (2005). Monocyte and macrophage heterogeneity. Nat Rev Immunol 5, 953-964.
- Harris JA et al. (2012). CD163 versus CD68 in tumor associated macrophages of classical Hodgkin lymphoma. Diagn Pathol 7, 12-17.
- Heusinkveld M and van der Burg SH (2011). Identification and manipulation of tumor associated macrophages in human cancers. J Transl Med 9, 216-229.
- Hu H et al. (2009). Tumor cell-microenvironment interaction models coupled with clinical validation reveal CCL2 and SNCG as two predictors of colorectal cancer hepatic metastasis. Clin Cancer Res 15, 5485-5493.
- Kitamura T et al. (2015). Immune cell promotion of metastasis. Nat Rev Immunol 15, 73-86.
- Komohara ND et al. (2010). Ratio of M2 macrophage expression is closely associated with poor prognosis for Angioimmunoblastic T-cell lymphoma (AITL). Pathol Int 60, 278-283.
- Komohara Y et al. (2014). Clinical significance of macrophage heterogeneity in human malignant tumors. Cancer Sci 105, 1-8.
- Komohara Y et al. (2015). Tumor-associated macrophages: Potential therapeutic targets for anti-cancer therapy. Adv Drug Deliv Rev doi: 10.1016/j.addr.2015.11.009. [Epub ahead of print].
- Krausgruber T et al. (2011). IRF5 promotes inflammatory macrophage polarization and TH1-TH17 responses. Nat Immunol 12, 231-238.
- Kryczek I et al. (2006). B7-H4 expression identifies a novel suppressive macrophage population in human ovarian carcinoma. J Exp Med 203, 871-881.
- Li SN et al. (2013). IL-21 modulates release of proinflammatory cytokines in LPS-stimulated macrophages through distinct signaling pathways. Mediators Inflamm 2013, 548073.
- Liao X et al. (2011). Krüppel-like factor 4 regulates macrophage polarization. J Clin Invest 121, 2736-2749.
- Mackaness GB (1962). Cellular resistance to infection. J Exp Med 116, 381-406.
- MacMicking J et al. (1997). Nitric oxide and macrophage function. Annu Rev Immunol. 15, 323-350.
- Mantovani A et al. (2004). The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol 25, 677-686.
- Mantovani A et al. (2006). Role of tumor-associated macrophages in tumor progression and invasion. Cancer Metastasis Rev 25, 315-322.
- Martinez FO and Gordon S (2014). The M1 and M2 paradigm of macrophage activation: time for reassessment. F1000Prime Rep 6, 1-13.
- Martinez FO et al. (2013). Genetic programs expressed in resting and IL-4 alternatively activated mouse and human macrophages: similarities and differences. Blood 121, e57-e69.
- Martinez-Pomares L and Gordon S (2012). CD169+ macrophages at the crossroads of antigen presentation. Trends Immunol 33, 66-70.
- Medrek C et al. (2012). The presence of tumor associated macrophages in tumor stroma as a prognostic marker for breast cancer patients. BMC cancer 12, 306-314.
- Mills CD et al. (2000). M-1/M-2 macrophages and the Th1/Th2 paradigm. J Immunol 164, 6166-6173.
- Mills CD et al. (2016). A breakthrough: macrophage-directed cancer immunotherapy. Cancer Res 76, 513-516.
- Mosser DM and Edwards JP (2008). Exploring the full spectrum of macrophage activation. Nat Rev Immunol 8, 958-969.
- Movahedi K et al. (2010). Different tumor microenvironments contain functionally distinct subsets of macrophages derived from Ly6C (high) monocytes. Cancer Res 70, 5728-5739.
- Nathan CF (2008). Metchnikoff’s legacy in 2008. Nature Immunol 9, 695-698.
- Nathan CF et al. (1983). Identification of interferon-gamma as the lymphokine that activates human macrophage oxidative metabolism and antimicrobial activity. J Exp Med 158, 670-689.
- Ni G et al. (2014). Btk regulates macrophage polarization in response to lipopolysaccharide. Plos One 9, e85834.
- Odegaard JI et al. (2007). Macrophage-specific PPARγ controls alternative activation and improves insulin resistance. Nature 447, 1116-1120.
- Oetke C et al. (2006). The antigen recognized by MOMA-1 is sialoadhesin. Immunol Lett 106, 96-98.
- Ohri CM et al. (2011). The tissue microlocalisation and cellular expression of CD163, VEGF, HLA-DR, iNOS, and MRP 8/14 is correlated to clinical outcome in NSCLC. Plos One 6, e21874.
- Porta C et al. (2009). Tolerance and M2 (alternative) macrophage polarization are related processes orchestrated by p50 nuclear factor κB. Proc Natl Acad Sci USA 106, 14978-14983.
- Puellmann K et al. (2006). A variable immunoreceptor in a subpopulation of human neutrophils. Proc Natl Acad Sci USA 103, 14441-14446.
- Qian BZ and Pollard JW (2010). Macrophage diversity enhances tumor progression and metastasis. Plos One 8, e84009.
- Rőszer T (2015). Understanding the mysterious M2 macrophage through activation markers and effector mechanisms. Mediators Inflamm 2015, 816460.
- Satoh T et al. (2010). The Jmjd3-Irf4 axis regulates M2 macrophage polarization and host responses against helminth infection. Nat Immunol 11, 936-944.
- Sica A and Bronte V (2007). Altered macrophage differentiation and immune dysfunction in tumor development. J Clin Invest 117, 1155-1166.
- Sica A and Mantovani A (2012). Macrophage plasticity and polarization: in vivo veritas. J Clin Invest 122, 787- 795.
- Sica A et al. (2006). Tumour-associated macrophages are a distinct M2 polarised population promoting tumour progression: potential targets of anti-cancer therapy. Eur J Cancer 42, 717-727.
- Sierra-Filardi E et al. (2011). Activin A skews macrophage polarization by promoting a proinflammatory phenotype and inhibiting the acquisition of anti-inflammatory macrophage markers. Blood 117, 5092-5101.
- Stein M et al. (1992). Interleukin 4 potently enhances murine macrophage mannose receptor activity: a marker of alternative immunologic macrophage activation. J Exp Med 176, 287-292.
- Takeda N et al. (2010). Differential activation and antagonistic function of HIF-α isoforms in macrophages are essential for NO homeostasis. Genes Dev 24, 491-501.
- Toshchakov V et al. (2002). TLR4, but not TLR2, mediates IFN-beta-induced STAT1alpha/beta-dependent gene expression in macrophages. Nat Immunol 3, 392-398.
- Wang R et al. (2011). Tumor-associated macrophages provide a suitable microenvironment for non-small lung cancer invasion and progression. Lung Cancer 74, 188-196.







