Article: High-Affinity Antibodies for Peptide Enrichment Immuno-MRM
-
Monoclonal Generation
-
Custom Recombinant Monoclonal Antibody Generation
-
Webinars, Videos and Technical Articles
- Webinar: Overcome the Challenges of PK Assay Development Using TrailBlazer Antibodies
- Webinar: Generation of SARS-CoV-2 antibodies in multiple formats within four weeks
- Webinar: Recombinant Antibodies with SpyTag Technology
- Webinar: Transform bioanalytical assays with TrailBlazer Antibodies
- Webinar: Control your critical antibody reagents and avoid assay failure
- Webinar: Improve your antibody drug development assays
- Webinar: Optimize your assays using recombinant antibodies selected for desired affinity
- Webinar: The making of recombinant anti-idiotypic antibodies for high performance in bioanalytical assays
- Webinar: Human recombinant antibodies as positive controls and calibrators
- Webinar: How to overcome assay challenges using custom recombinant antibodies
- Webinar: Generation of high affinity recombinant antibodies for application in immuno-MRM
- Video: Generating anti-idiotypic antibodies for bioanalytical assays
- Video: Best practices for characterization and QC of anti-idiotypic antibodies for bioanalysis
- Video: Generation of drug-target complex specific antibodies
- Video: Antibodies for CAR-T cell therapy development
- Article: Monitoring antibody immune responses against biotherapeutic drugs
- Article: Effective tools for drug monitoring assays
- Article: An accelerated approach to sensitive ADA assays
- Article: Isolation of enzyme active site-specific recombinant antibodies by guided selection
- Article: Biomarker Assay Development using Highly Specific Recombinant Antibodies
- Article: Recombinant antibodies as standards for immunodiagnostic assays
- Article: High affinity antibodies for peptide enrichment immuno-MRM
- Article: Generation of antibodies against self-antigens
- Article: Generation and characterization of drug-target complex-specific antibodies
- Article: Antibodies for CAR-T Cell Therapy Development
- Poster: Faster Generation of Anti-Drug Antibodies Using SpyTag Technology
- Poster: CAR T cell analysis with modular antibodies
- Poster: Drug-Target-Complex Specific Antibodies for Pharmacokinetic Analysis of Biotherapeutics
- Poster: Characterization of anti-idiotypic antibodies for high performance in bioanalytical assays
- Poster: Recombinant anti-idiotypic antibodies for antibody drug development
- Poster: Generation of recombinant antibodies for Bio-Plex assays
-
Webinars, Videos and Technical Articles
-
Custom Recombinant Monoclonal Antibody Generation
s
Simplified sourcing via Scientist.com
s
Custom antibody project inquiry form
A personal, no obligation quotation for a custom monoclonal antibody generation project
s
Contact our custom antibody specialists
Tel: +49 (0) 89 80 90 95 45
Fax: +49 (0) 89 80 90 95 50
Office: Bio-Rad AbD Serotec GmbH, Campus Neuried, Anna-Sigmund-Str. 5, 82061 Neuried, Germany
Multiple reaction monitoring-mass spectrometry (MRM-MS) is an emerging technology for the quantitative analysis of peptide and protein expression with high precision and specificity. Such analysis of protein targets of interest has multiple applications; for example, in discovery research to look at changing levels of proteins upon disruption of a protein network, or in clinical research to detect proteins in plasma and tissue to verify and monitor human disease biomarker candidates. This technical article summarizes a feasibility study to discover whether recombinant anti-peptide antibodies generated using the HuCAL® antibody library and phage display technology are suitable for immunoaffinity enrichment of peptides coupled to targeted MRM.

Generation of High-Affinity Recombinant Antibody Fragments for Peptide Enrichment Immuno-MRM Assays
Multiple reaction monitoring (MRM) is a method of protein quantitation that obtains precise and sensitive measurements of specific target analytes in complex biological samples. Prior to performing MRM, many peptide analytes require an enrichment step to achieve sufficient sensitivity in the assay. Coupling a peptide immunoaffinity enrichment step to MRM (i.e. an immuno-MRM assay) is effective for most analytes and provides high sensitivity and specificity, a wide dynamic range, and convenience of sample handling when measuring endogenous proteins. Furthermore, immuno-MRM is reproducible across laboratories and capable of multiplexing several analytes simultaneously.
Immuno-MRM using recombinant Fab antibodies
An increasing number of studies (such as those by Schoenherr et al. 2010 and Razavi et al. 2011) have investigated immuno-MRM using monoclonal antibodies. The renewable nature, specificity profile, and uniform affinity of monoclonal antibodies make them preferable to affinity-purified polyclonal antibodies for immuno-MRM. However, the great expense and amount of time required for large-scale generation of hybridoma-based monoclonal antibodies has limited the widespread application of immuno-MRM.
High-affinity recombinant antibody fragments (Fabs) isolated from large naïve libraries may offer several advantages over both polyclonal and monoclonal antibodies. The in vitro generation of Fabs in E. coli and other bacteria allows the user to control selection parameters (including affinity), access a wider range of antigens (including toxic, conserved or self-antigens), automate the process, and complete the procedure more rapidly. However, no previous studies have investigated whether 1) high-affinity recombinant antibodies against short linear peptides can be selected from a naïve library or 2) the recombinant antibodies obtained as fragments expressed from E. coli are suitable for procedures involving immunocapture of small linear peptides.
Therefore, Whiteaker and colleagues (Whiteaker et al. 2014) at the Fred Hutchinson Cancer Research Center and Bio-Rad described the generation, expression, and characterization of monovalent Fabs and determined the feasibility of using these Fabs in peptide immuno-MRM assays. Mouse osteopontin, human E-selectin, and human ADAM17 were selected because they all had peptide antigens with high-affinity monoclonal benchmark antibodies that had been generated previously by traditional immunization. The target peptide sequences were proteotypic for each of the respective proteins. Full-length IgG antibodies were also created from these recombinant antibodies, and the full-length IgG derivatives and recombinant Fab antibodies were compared to traditional anti-peptide rabbit or mouse monoclonal antibodies.
Rapid affinity maturation for generation of anti-peptide antibodies
The HuCAL PLATINUM® library (Prassler et al. 2011) along with RapMAT® technology (Prassler et al. 2009) and inhibition screening with free peptide were used to generate recombinant antibodies rapidly with a high affinity to the free peptides. Phage display selection (i.e. panning) involved immobilization of the antigen on magnetic beads, incubation with the HuCAL library, washing steps to remove unspecific antibodies and elution of the phage encoding the enriched antibodies (Figure 1). Two rounds of panning on the peptide-transferrin and bovine serum albumin conjugates, respectively, were used. After the first two rounds, libraries for affinity maturation were generated. This process involved the exchange of the gene region coding for the CDR3s of the antibody light chains (LCDR3) from the identified pool of Fab genes, with a highly diverse LCDR3 maturation cassette.
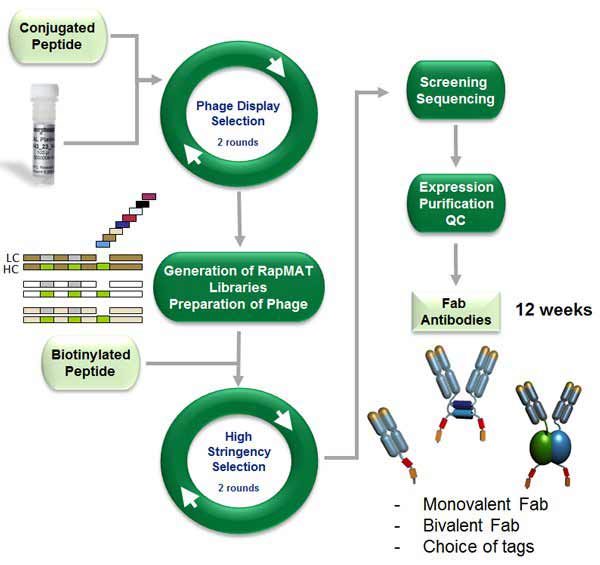
Fig. 1. Scheme of the antibody generation process with the HuCAL phage display antibody library and RapMAT technology.
The Fab displaying phage from the new antibody libraries went through another two rounds of panning at higher stringency, and using two different strategies - either decreasing amounts of antigen-coupled magnetic beads to capture peptide carrier protein conjugates or streptavidin-coated beads to capture biotinylated peptides. The biotinylated peptides captured by streptavidin beads selected significantly more antibodies capable of binding to the free peptide in solution than did the peptide carrier protein conjugate-coupled magnetic beads system.
After panning, the pool of Fab genes was subcloned into an E. coli expression vector, leading to functional periplasmic expression of monovalent Fabs that were used for affinity purification and capture in immuno-MRM experiments. For each antigen, the two Fabs with the highest affinity for the free peptide were converted to the hIgG1 format and produced in mammalian cell culture. To initially assess the performance of the Fabs and hIgG1 products of the six clones, peptide recoveries were estimated by measuring the amount of spiked light peptide relative to heavy peptide (a stable isotope-labeled standard) before and after the enrichment process. For the peptides corresponding to osteopontin and E-selectin, recoveries were greater than 85% for the Fabs and hIgG1 antibodies and at least as high as those of the corresponding rabbit monoclonal antibodies. Furthermore, generation of recombinant Fab antibodies and subsequent conversion to full-length IgG format (including production and quality control) took about 20 weeks in this study, whereas generation of animal-derived monoclonal antibodies typically takes 6-9 months. Use of streptactin-coated beads allowed direct use of the Strep tagged Fabs in the immuno-MRM assay and eliminated the need for additional steps to convert the Fabs to full-length immunoglobulins, reducing the antibody generation timeline to 12 weeks. Moreover, the process is highly automated and as a consequence, many antibody generation projects can be run in parallel without a corresponding increase in timelines. Therefore, use of recombinant Fabs equipped with affinity tag for capture to beads could allow for efficient and simultaneous completion of multiple projects. Even though the researchers successfully generated Fabs that targeted the peptide sequences for their proteins of interest, some peptides may not yield high-affinity recombinant antibodies. Further studies are needed to investigate the use of diverse targets for properties, such as peptide length, hydrophobicity, charge, and post-translational modifications, when developing high-affinity recombinant antibodies.
Fab antibodies successful in immuno-capture experiments
To assess the performance of the reagents in the immuno-MRM assay, the researchers measured the linear range, limit of detection (LOD), and precision for response curves of each peptide/antibody combination suspended in a matrix containing denatured, digested human plasma (adjusted to pH 8.0 before addition of antibodies), antibodies, variable amounts of isotope-labeled heavy synthetic peptides, a constant amount of light synthetic peptides, and magnetic beads mixed in PBS containing 0.03% CHAPS. The overall dynamic range of response was at least three to four orders of magnitude, and with the exception of the full length IgG antibody for ADAM 17, the response curves for the Fab, full-length IgG, and monoclonal antibody were nearly identical for each of the respective peptides (Figure 2). The LODs and precisions (measured as a percent coefficient of variation) were comparable among all of the reagents, although the converted full-length IgG antibody for the ADAM17 peptide only detected the peptide at the highest concentration. Nevertheless, the best Fab for the ADAM17 peptide was still successful in immuno-capture experiments even though only medium-affinity antibodies were isolated. According to the researchers, this suggests that antibodies directly selected from the HuCAL library could be used in the immuno-MRM assays without employing preliminary affinity maturation steps, which would further shorten the timeline for Fab antibody generation from 12 to 8 weeks.
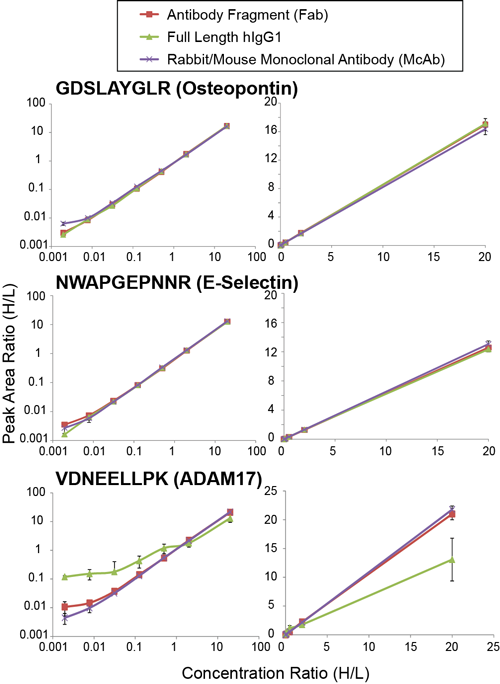
Fig. 2. Response curves for each peptide/affinity reagent. The concentration of heavy peptide was varied and measured relative to the light peptide signal. One antibody fragment and full length IgG was used for each peptide, GDSLAYGLR (AbD18303), NWAPGEPNNR (AbD18279), and VDNEELLPK (AbD18307). For each peptide, the curve is plotted on a log10 scale and a linear scale. Curves obtained from using antibody fragments (red), IgG (green), and rabbit or mouse monoclonal antibodies (purple) are overlaid. Error bars are the standard deviation of three replicate measurements.
In conclusion, use of isolated antibody fragments from a naïve antibody library is feasible in immuno-MRM-based quantitation. Additionally, use of recombinant antibodies generated in vitro allows projects to be completed in a relatively short time frame by taking advantage of an automated procedure and controlling selection parameters such as affinity. Furthermore, the sequence of the recombinant antibody is known and the genes are available on plasmids, which enables conversion of the Fabs to full immunoglobulins or addition of peptide tags and/or multimerization domains. To determine the large-scale success rate of this approach, further research will be needed using a larger and more diverse set of analytes.
References
-
Prassler et al (2009). In vitro affinity maturation of HuCAL antibodies: complementarity determining region exchange and RapMAT technology. Immunotherapy 4, 571-583.
- Prassler et al (2011). HuCAL PLATINUM, a synthetic Fab library optimized for sequence diversity and superior performance in mammalian expression systems. J Mol Biol 1, 261-278.
- Razavi M et al (2011). MALDI immunoscreening (MiSCREEN): a method for selection of anti-peptide monoclonal antibodies for use in immunoproteomics. J Immunol Methods 1-2, 50-64.
- Schoenherr RM et al (2010). Automated screening of monoclonal antibodies for SISCAPA assays using a magnetic bead processor and liquid chromatography-selected reaction monitoring mass spectrometry. J Immunol Methods 1-2, 49-61.
- Whiteaker et al (2014). High-affinity Recombinant Antibody Fragments (Fabs) Can Be Applied in Peptide Enrichment Immuno-MRM Assays. J Proteome Res 13, 2187-2196.
Figures 1 and 2 are reproduced with permission from The Journal of Proteome Research, Figures 1 and 3 respectively, http://pubs.acs.org/doi/pdf/10.1021/pr4009404 ; Figure 1 has been modified to include bivalent Fab antibody formats.





