Article: Isolation of Enzyme Active Site-Specific Recombinant Antibodies by Guided Selection
-
Monoclonal Generation
-
Custom Recombinant Monoclonal Antibody Generation
-
Webinars, Videos and Technical Articles
- Webinar: Overcome the Challenges of PK Assay Development Using TrailBlazer Antibodies
- Webinar: Generation of SARS-CoV-2 antibodies in multiple formats within four weeks
- Webinar: Recombinant Antibodies with SpyTag Technology
- Webinar: Transform bioanalytical assays with TrailBlazer Antibodies
- Webinar: Control your critical antibody reagents and avoid assay failure
- Webinar: Improve your antibody drug development assays
- Webinar: Optimize your assays using recombinant antibodies selected for desired affinity
- Webinar: The making of recombinant anti-idiotypic antibodies for high performance in bioanalytical assays
- Webinar: Human recombinant antibodies as positive controls and calibrators
- Webinar: How to overcome assay challenges using custom recombinant antibodies
- Webinar: Generation of high affinity recombinant antibodies for application in immuno-MRM
- Video: Generating anti-idiotypic antibodies for bioanalytical assays
- Video: Best practices for characterization and QC of anti-idiotypic antibodies for bioanalysis
- Video: Generation of drug-target complex specific antibodies
- Video: Antibodies for CAR-T cell therapy development
- Article: Monitoring antibody immune responses against biotherapeutic drugs
- Article: Effective tools for drug monitoring assays
- Article: An accelerated approach to sensitive ADA assays
- Article: Isolation of enzyme active site-specific recombinant antibodies by guided selection
- Article: Biomarker Assay Development using Highly Specific Recombinant Antibodies
- Article: Recombinant antibodies as standards for immunodiagnostic assays
- Article: High affinity antibodies for peptide enrichment immuno-MRM
- Article: Generation of antibodies against self-antigens
- Article: Generation and characterization of drug-target complex-specific antibodies
- Article: Antibodies for CAR-T Cell Therapy Development
- Poster: Faster Generation of Anti-Drug Antibodies Using SpyTag Technology
- Poster: CAR T cell analysis with modular antibodies
- Poster: Drug-Target-Complex Specific Antibodies for Pharmacokinetic Analysis of Biotherapeutics
- Poster: Characterization of anti-idiotypic antibodies for high performance in bioanalytical assays
- Poster: Recombinant anti-idiotypic antibodies for antibody drug development
- Poster: Generation of recombinant antibodies for Bio-Plex assays
-
Webinars, Videos and Technical Articles
-
Custom Recombinant Monoclonal Antibody Generation
s
Simplified sourcing via Scientist.com
s
Custom antibody project inquiry form
A personal, no obligation quotation for a custom monoclonal antibody generation project
s
Contact our custom antibody specialists
Tel: +49 (0) 89 80 90 95 45
Fax: +49 (0) 89 80 90 95 50
Office: Bio-Rad AbD Serotec GmbH, Campus Neuried, Anna-Sigmund-Str. 5, 82061 Neuried, Germany
Human Combinatorial Antibody Library (HuCAL®) Technology Used to Isolate Antibodies Specific to the Active Site of the West Nile Virus NS2B-NS3 Proteinase Complex
Selection of antibodies from the antibody library is performed in vitro. This enables greater flexibility for antibody generation than is available with conventional methods based on the immunization of animals. Excellent antibody specificity can be achieved using ‘Guided Selection’. In this example, antibodies specific to the West Nile Virus (WNV) NS2B-NS3 proteinase active site were isolated using phage display blocking strategies, providing antibody research tools that enabled scientists to carry out inhibition experiments and structural studies.
In vitro characterization of enzymatic function and experiments to measure the dynamics of substrate binding and catalysis typically involve the use of inhibitory agents. Unfortunately, small molecule inhibitors that block the activity of an enzyme with high specificity are often not available. Indeed, many inhibition studies are a necessary precursor to the development of, or screening for, therapeutic small molecule inhibitors.
Antibodies or their Fab domain fragments make ideal probes and inhibitors for these early stage functional studies. They can bind to target enzymes with high affinity and specificity, for example differentiating between phosphorylated and non-phosphorylated proteins or mutant and wild-type enzymes.
A team of researchers, led by Dr Alex Strongin from the Sanford Burnham Prebys Medical Discovery Institute (USA), worked with scientists from Bio-Rad to isolate high affinity antibodies from the synthetic naïve Human Combinatorial Antibody Library (HuCAL) that were specific to the active site of West Nile Virus NS2B-NS3 proteinase (Shiryaev et al. 2010).
West Nile Virus (WNV) is a mosquito-borne flavivirus. Most infections are asymptomatic; less than 1% of infected people develop serious neuroinvasive illness that can be fatal. Cases of WNV are on the rise, especially in the USA. Over 2,200 cases were reported to the Centers for Disease Control and Prevention in 2014, including almost 100 deaths.
No treatment or vaccine is available yet, although an investigational hydrogen peroxide-based vaccine entered phase I human trials in July 2015. The virus genomic RNA codes for 10 proteins: three structural proteins and seven non-structural proteins or enzymes. These proteins are produced by processing a single precursor polypeptide. NS2B-NS3 is the only proteinase in the WNV genome; it is involved in essential post-translational processing of viral proteins and could be a suitable target for small molecule WNV treatments.
The HuCAL PLATINUM® library contains more than 45 billion human antibodies against virtually every possible antigen or epitope, from post-translational modifications to active site in an enzyme
About HuCAL Technology
The HuCAL library has been created by mirroring the structural repertoire of the human immune system through the generation of master genes representing the various subclasses of human immunoglobulin (Knappik et al. 2000). Sequences for the highly diversified complementarity determining regions (CDRs) of the antibody binding sites are synthesized by using trinucleotides. These ‘cassettes’ are combined to create antibody genes that code for some 45 billion unique fully human antibodies. The HuCAL library is the largest, fully synthetic, fully human antibody library available today, and the technology is well published. It has been used by the Bio-Rad custom antibody team to generate antibodies for research and diagnostic applications since 2004. Screening of the HuCAL antibody library is performed in vitro, which allows the selection of highly specific antibodies that recognize a distinct epitope, such as the active site of an enzyme. Such specificity is very difficult to achieve using traditional monoclonal antibody generation methods that rely on the immunization of animals.
As the sequence of any selected antibody is known, it is possible to reproduce the genes synthetically if needed. This sequence back up secures the future supply of the antibody and recombinant production methods ensure there is a high level of consistency between batches.
Guided Selection Strategies to Select Highly Specific Antibodies
The Bio-Rad team ‘panned’ the library for antibodies with specificity for active-site binding. They used variations of a technique called phage display blocking to identify and select phages and isolate the genes for antibodies with active site specificity for the WNV NS2B-NS3 proteinase complex.
The phage library was blocked with a mutant NS2B-NS3 construct (H51A) in which a histidine residue had been replaced by alanine within the NS2B-NS3 active site, Figure 1. This substitution mutation only altered the active site, keeping all other putative epitopes of the wild-type enzyme intact. Hence the mutant could be used to block members of the antibody library that bind to epitopes outside the active site, Figure 2.
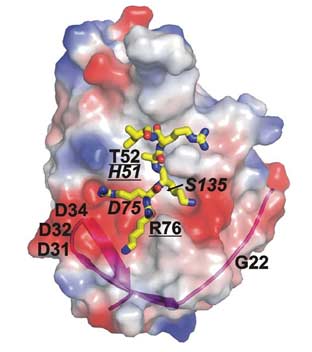
Fig. 1. The structure of WNV NS2B-NS3pro. The original position of the His51, Asp75 and Ser135 of the catalytic triad (italicized) and the mutant positions are shown. The mutations that affect the binding of the proteinase with the antibodies are underlined. The surface is colored by electrostatic potential (negative, red; positive, blue). The magenta ribbon shows NS2B contributing to the NS3pro active site. The positioning of a substrate-mimetic peptide (KRKARI) is shown in yellow. The model was presented using PyMol Software (DeLano Scientific).
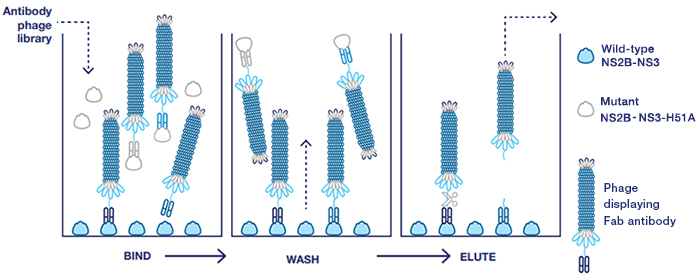
Fig. 2. Guided selection with blocking. The mutant NS2B-NS3 construct (H51A) was used to remove antibodies binding to epitopes outside the active site (counter selection).
The rationale was that phage-displayed antibodies recognizing NS2B-NS3 proteinase outside of the mutant active site were unlikely to bind to the wild-type NS2B-NS3 protein antigen immobilized in the wells (in reality the antigen was a slightly altered mutant to prevent autocleavage of the enzyme). The only phages that would bind to the immobilized wild-type proteinase were those with antibody specific to the wild-type active site.
Unbound and blocked phages were removed by washing; the bound phages were eluted for further rounds of panning under the same stringent blocking conditions to ensure that only phages displaying antibodies that recognize the wild-type active site were isolated for sequencing and later production in E. coli, Figure 3.
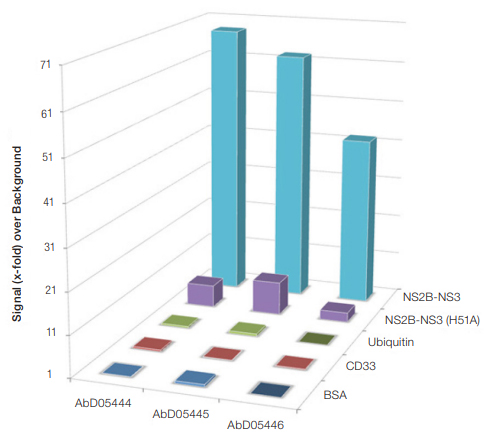
Fig. 3. Indirect ELISA. Data showing three of the antibodies selected from the phage library that bind to the wild-type enzyme active site (NS2B-NS3) and not the inert H51A mutant. BSA, ubiquitin and CD33 were used as controls to eliminate false-positive antibodies.
Altogether seven antibodies were obtained with this strategy, and four of them were later shown to inhibit the enzymatic activity of NS2B-NS3; data for three of these is shown in Figure 4.
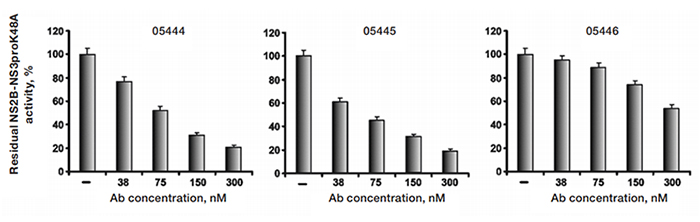
Fig. 4. Selected antibodies efficiently inhibit the catalytic activity of WNV NS2B-NS3. Before the addition of the Pyr-RTKR-AMC substrate (25 μM), the purified WNV proteinase (10 nM) was co-incubated for 30 minutes with increasing concentrations of the antibodies. The residual activity was then monitored continuously at λex =360 nm and λem =465 nm to determine the initial velocity of the reactions. The untreated proteinase was used as a control and its activity was counted as 100%.−, no antibody; Ab, antibody.
Conclusion
Phage display selection on the NS2B-NS3 proteinase under conditions where a single-point active-site mutant of the enzyme was present in excess amounts led to the isolation of antibodies that were specific to the functional active site of wild-type WNV NS2B-NS3 proteinase. The Strongin team demonstrated the selective active site binding of the antibodies in substrate-cleavage reactions (those with bound antibody were unable to function) and additional screens with other active site single mutations and NS2B-NS3 proteinases from Dengue virus type 2, hepatitis C virus or human serine proteinase. The antibodies did not bind to any of these homologous enzymes. Following this feasibility study, the team has also used the antibodies for mutagenesis and structure-based optimization and as experimental NS2B-NS3 probes. Structural studies of the antibody-proteinase active site binding and dynamics could help to guide the design of selective small molecule flaviviral proteinase inhibitors.
As demonstrated in this study, antibodies that are highly specific to an enzyme active site can be successfully isolated from the HuCAL antibody library using in vitro guided selection strategies. These antibodies provide inhibitory reagents that enable researchers to characterize enzymatic function and measure the dynamics of substrate binding and catalysis, which could contribute to advances in therapeutic or vaccine development.
References
- Knappik A et al. (2000). Fully synthetic human combinatorial antibody libraries (HuCAL) based on modular consensus frameworks and CDRs randomized with trinucleotides. J Mol Biol. 296:57-86
- Shiryaev S et al. (2010). Isolation and characterization of selective and potent human Fab inhibitors directed to the active-site region of the two-component NS2B–NS3 proteinase of West Nile virus. Biochem J. 427:369-376
Figures 1, 3 and 4 are reproduced with permission of Portland Press from Shiryaev et al (2010). Figures have been modified from the original as follows: Figure 1 shows only the 3D structure, Figure 3 is shown in color, Figure 4 shows data for only three out of five antibodies.






