Ubiquitination Mediated Signaling

- On This Page
- Antibodies for the analysis of ubiquitination
- Ubiquitination antibodies
- References
s
Western Blot University
s
What are R-loops?
s
Cancer Pathway Posters
Compiling key proteins involved in key signaling pathways: cGAS-STING, EGF R, mTOR, NF-kB, and PI3/AKT pathways.
s
Recent Blog
Antibodies for the Analysis of Ubiquitination
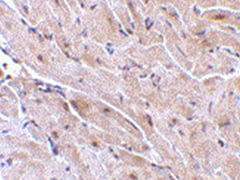
Staining of formalin fixed, paraffin embedded mouse heart stained with rabbit anti TRIAD3A antibody (AHP1000) following antigen retrieval using sodium citrate buffer (pH 6.0).
Related blogs
The emerging importance of ubiquitination regulators in cancer therapy
SUMOylation: a Heavyweight in the Regulation of Cell Cycle Progression and Cell Proliferation
Ubiquitination, the conjugation of ubiquitin moieties to lysine residues, functions as a signal for targeting proteins for degradation by the 26S proteasome. However, in addition to this now “classical” role of ubiquitination, other effects of this post-translational modification, such as the mediation of DNA damage responses and membrane trafficking have been discovered (Nathan et al. 2013).
The impact of an ubiquitination event depends primarily on whether a single ubiquitin moiety (monoubiquitination) or an ubiquitin chain (polyubiquitination) has been conjugated to a substrate. Monoubiquitination, especially that of receptor tyrosine kinases (RTKs) such as the epidermal growth factor receptor, results in RTK internalization followed by degradation via the endosomal/lysosomal pathway (Haglund et al. 2012).
The impact of polyubiquitination differs depending on how the attached ubiquitin chain has been linked. Ubiquitin contains seven lysine residues (K6, K11, K27, K29, K33, and K48), which all provide a basis for the formation of an ubiquitin chain (Nathan et al. 2013). K11- and K48-linked ubiquitin chains have been reported as signals for proteasomal degradation (Wickliffe et al. 2011). K63-linked chains, on the other hand, have been shown to play a role in the NF-kappa B signaling pathway, DNA damage response and to function as signals for the trafficking of proteins to lysosomes (Nathan et al. 2013, Walczak et al. 2012).
The ubiquitination process, independent of whether a protein is mono- or polyubiquitinated, is orchestrated by three different enzyme classes known as E1 (ubiquitin-activating enzyme), E2 (ubiquitin-conjugating enzyme) and E3 (ubiquitin ligase) enzymes (Walczak et al. 2012). Once conjugated, ubiquitin moieties can be removed from protein substrates by DUBs (deubiquitination enzymes) (Coyne and Wing 2016).
Bio-Rad offers a variety of antibodies for the detection of ubiquitination and the ubiquitin cascade components by ELISA, immunofluorescence, immunohistochemistry, immunoprecipitation and western blotting.
Antibodies for the Analysis of Ubiquitination
| Description | Target | Format | Clone | Applications | Citations | Code |
|---|
- Growth factors
- EGF R resources
- Kinases
- mTOR signaling pathway
- Transcription factors
- Translation factors
- Phosphatases
- Phosphatases in cancer
- Phospho-Specific precisionAb antibodies
- Phospho-specific antibodies
- Sumoylation mediated signaling
- Tyrosine phosphorylation of EGF R
- Signal transduction antibodies
References
- Coyne ES and Wing SS (2016). The business of deubiquitination – location, location, location. F1000Res 11, 5.
- Haglund K and Dikic I (2012). The role of ubiquitylation in receptor endocytosis and endosomal sorting. J Cell Sci 125, 265-275.
- Nathan JA et al. (2013). Why do cellular proteins linked to K63-polyubiquitin chains not associate with proteasomes? Embo J. 32, 552-565.
- Walczak H et al. (2012). Generation and physiological roles of linear ubiquitin chains. BMC Biology 10:23.
- Wickliffe KE et al. (2011). K11-linked ubiquitin chains as novel regulators of cell division. Trends Cell Biol 21, 656-663.