Immune Response to Influenza A Virus
Influenza viruses are respiratory negative-stranded RNA viruses from the Orthomyxoviridae family causing an acute febrile disease referred to as influenza or more commonly — flu. Influenza viruses exist as four types (A, B, C, and D); type A can infect a wide variety of species, type B only affects humans, type C infects humans and pigs, and type D is specific for cattle and swine however, antibodies to type D viruses have been found in humans (Trombetta et al. 2019).
Influenza Virus Structure, Types, and Strains
A key feature of influenza viruses is the antigenic drift and reassortment that makes them unpredictable, necessitating seasonal changes to the flu vaccine. While both Influenza A and B viruses cause seasonal flu epidemics, influenza B typically emerges later in the season and causes up to a third of seasonal flu infections (Dumm and Heaton 2019), while influenza A virus often leads to pandemics.
Influenza A
The influenza A genome is made up of eight segments coding for 11 viral proteins (Table 1).
Table 1. RNA segments of influenza A viruses.
| Segment | Protein(s) | Protein function | |
|---|---|---|---|
| 1 | PB2 | Polymerase basic 2 | Polymerase subunit; mRNA cap recognition |
| 2 | PB1 | Polymerase basic 1 | Polymerase subunit; RNA elongation, endonuclease activity |
| PB1-F2 | Polymerase basic 1 — 2nd ORF | Pro-apoptotic activity | |
| 3 | PA | Polymerase acidic | Polymerase subunit; protease activity |
| PA-X | Polymerase acidic – frameshift | Polymerase subunit; suppresses accumulation of host mRNAs | |
| 4 | HA | hemagglutinin | Surface glycoprotein; major antigen, receptor binding, and fusion activities |
| 5 | NP | Nucleoprotein | RNA binding protein; nuclear import regulation |
| 6 | NA | Neuraminidase | Surface glycoprotein; sialidase activity, virus release |
| 7 | M1 | Matrix protein 1 | Matrix protein; vRNP interaction, RNA nuclear export regulation, viral budding |
| M2 | Matrix protein 2 – splice variant | Ion channel; virus uncoating and assembly | |
| 8 | NS1 | Non-structural protein 1 | Interferon antagonist protein; regulation of host gene expression |
| NS2 (NEP) | Non-structural protein 2–splice variant | Nuclear export of RNA | |
Modified from Table 1 in (Bouvier and Palese 2008).
The viral envelope contains the transmembrane proteins HA, NA, and M2. The most virulent types of influenza A viruses are mainly referred to by their subtypes with this classification based on the expression of two surface proteins — HA and NA with 18 HA and 11 NA subtypes in existence. Among these subtypes, several have become familiar names even with the general population due to their association with disease outbreaks. The H5N1 strain (often referred to as bird flu) originated in 2003 from avian species while the H1N1 strain (often called swine flu) emerged in 2009 and is derived from swine (Sims et al. 2005, Vijaykrishna et al. 2010).
Influenza B, C, and D
Instead of being classified by subtypes, influenza B viruses are classified into two strains — B/Yamagata and B/Victoria. To date, influenza type C and D viruses have not been assigned strains or subtypes.
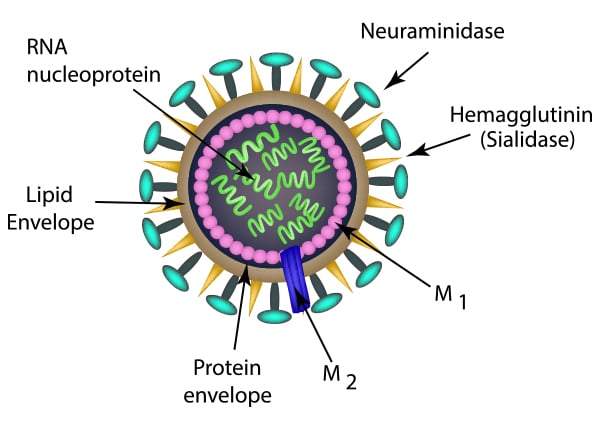
Fig. 1. The structure of the influenza virion.
Virus-Host Cell Interaction
The influenza virus replicates by infecting the bronchiolar-alveolar epithelial cells in the respiratory tract, and spreads throughout the airways from the initial infection site. To establish an infection, the influenza virus needs to first overcome host immune responses, including the mucosal barrier to access the underlying epithelium. Different strains of influenza A virus make use of different mechanisms to enter the host via the upper respiratory tract. The H1N1 subtype binds to ciliated epithelial cells and goblet cells, whereas H5N1 rarely attaches to these cells in humans (van Riel et al. 2010). Instead, H5N1 favors infection via alveolar macrophages and alveolar epithelial cells (van Riel et al. 2006, Nicholls et al. 2007). Similarly, infection via the lower respiratory tract utilizes different mechanisms depending on the strain involved, for example, H1N1 and H3N2 are connected to the human trachea and bronchi more than H5N1 (van Riel et al. 2007).
Sialic acid moieties found in the mucin glycoproteins MUC5AC, MUC5B, and MUC1 in the host respiratory mucus layer act as an important defense mechanism for slowing down influenza A virus infection (Ehre et al. 2012, McAuley et al. 2017). The mucin-associated sialic acid acts as a decoy receptor for influenza A HA; however, this defense is countered by the NA enzyme that breaks it down (Cohen et al. 2013).
Ultimately though, HA will attach to the cell via α2,3 and α2,6-linked sialic acid of cell surface N-glycans. Avian influenza A viruses binding to the former, and human ones to the latter. More recent work suggests that O-glycans and glycosphingolipid (GSL)-glycans and even non-sialylated, phosphorylated glycans are also involved in the initial binding of the influenza A virus.
This attachment phase is followed by the binding of the internalization receptors. Several molecules have been suggested to be the internalization receptors, among them, nucleolin (Chan et al. 2016), epidermal growth factor receptor (EGFR) (Eierhoff et al. 2010), and voltage-dependent calcium channel CaV1.2 (Fujioka et al. 2018). A sialic acid independent internalization can be carried out by C-type lectins (Upham et al. 2010, Ng et al. 2014). DC-SIGN (DC209) and L-SIGN (CD209L) have been shown to facilitate the take up of influenza A virus (Londrigan et al. 2011).
Cell Entry
Virus attachment to the host cell and binding by the internalization receptor induces clathrin-dependent endocytosis involving dynamin (Roy et al. 2000, Rust et al. 2004). While clathrin interacts with several proteins to induce membrane budding, dynamin drives the last stage of detachment from the cell membrane and endocytic vesicle formation. The less specific and receptor-independent macropinocytosis is an alternative entry pathway for the influenza A virus (de Vries et al. 2011). In either pathway, the endocytic vesicles fuse with the endosomes where the acidic environment of the endosome causes the opening of the M2 proton channels, inducing a conformational change in HA, and exposing the fusion peptide of the HA2 subunit needed for membrane fusion (Li et al. 2014). After viral membrane fusion with the endosome, the viral ribonucleoproteins (vRNPs) are released from the M1 protein, freeing them to diffuse into the host cell’s cytosol (Fontana et al. 2012). The vRNPs are then translocated into the host nucleus to initiate the pathway that leads to the production of viral proteins and RNA (Samji 2009).
In the nucleus, the RNA-dependent RNA polymerase (RdRP), consisting of PB1, PB2, and PA, carries out transcription and replication of the viral RNA genome. PB2 binds the 5′ cap of host pre-mRNA, and PA cuts host mRNA to supply 5′-capped RNA primer fragments to PB1 which then initiates viral mRNA transcription. In contrast, viral RNA replication does not need to be primed by 5′ caps of host pre-mRNA. (Dias et al. 2009, Pflug et al. 2017). Once the viral RNA has been transcribed, the host’s ribosomes synthesize the viral proteins. The viral polymerases reenter the nucleus where they are complexed with the M1 protein and newly synthesized viral RNA, ready for subsequent nuclear export facilitated by the NS2 protein. The NA, HA, and M2 proteins are processed via the endoplasmic reticulum through the Golgi to the host cell plasma membrane. They are then joined by the viral ribonucleoproteins arriving from the nucleus to form new virions that will bud off from the host cell membrane.
Innate Immune Response
Pattern Recognition Receptors Response to Influenza A
After the mucins in the airways, the next immune defense the influenza virus encounters is the pattern recognition receptors (PRRs) which bind pathogen-associated molecular patterns (PAMPs). PRRs are expressed on and inside immune cells and also epithelial cells, where they recognize PAMPS and trigger an immune response. PRRs involved in the recognition of influenza PAMPs are all intracellular, among them, Toll-like receptors (TLRs), nucleotide-binding oligomerization domain (NOD)-like receptor family pyrin domain-containing 3 (NLRP3), and retinoic acid-inducible gene I (RIG-I). Triggering of these PRRs drives the expression and secretion of proinflammatory cytokines and type I interferons (IFNs) which enhance antiviral innate immune responses. Cyclic guanosine monophosphate-adenosine monophosphate synthase (cGAS), while a cytosolic DNA sensing molecule, is also involved in the defense against RNA viruses such as influenza A. It also triggers the release of type IFNs (Schoggins et al. 2014, Sun et al. 2013, Webb and Fernandez-Sesma 2022).
TLR3
TLR3 is located in endosomes and lysosomes and recognizes double-stranded RNA (dsRNA). The involvement of TLR3 in the immune defense against influenza viruses is not fully elucidated yet, as the specific viral RNA structure bound by TLR3 has not been identified. However, influenza virus-infected human respiratory epithelial cells which constitutively express TLR3, drive a pro-inflammatory cytokine response (Le Goffic et al. 2007). Additionally, influenza infection in TLR3 −/− mice leads to a higher viral load but avoids hyperinflammatory damage, supporting a role for TLR3 (Le Goffic et al. 2006). Further evidence has shown that a single-stranded oligonucleotide can inhibit TLR3 activation in influenza A infection models (Poux et al. 2019). All supporting a role for TLR3 in anti-influenza A infection.
TLR7
TLR7, also located in the endosomes of endothelial and innate immune cells such as alveolar macrophages, dendritic cells (DCs), and neutrophils, binds influenza ssRNA as the virus is transported in the endosomes. This binding will induce type I IFNs and pro-inflammatory cytokines secretion by signaling through MyD88, IFN-regulatory factor 7 (IRF7), and activating nuclear factor-κB (NF-κB).
RIG-I
Beyond the endosomes, cytosolic receptors such as RIG-I are the next immune defense. RIG-I detects short viral 5′-triphosphorylated ssRNA or dsRNA in infected epithelial cells, conventional DCs (CDCs), and alveolar macrophages (Kato et al. 2005). RIG-I senses the viral RNA in anti-viral stress granules, a site where viral RNA and IFNg stimulated gene products, such as the serine/threonine kinase protein kinase R (EIF2AK2) accumulate (Onomoto et al. 2012). RIG-I induces the mitochondrial antiviral signaling protein (MAVS, IPS-1) resulting in the activation of IRF3/7 and NF-κB, which in turn then initiate the production of proinflammatory cytokines and type I IFN.
NLRP3
The NLR family pyrin domain-containing protein 3 (NLRP3) is found in monocytes, DCs, neutrophils, and macrophages, and also in human bronchial epithelial cells (Pothlichet et al. 2013). NLRPs assemble into multiprotein inflammasome complexes made up of an NLRP (or other PRRs), the adaptor ASC and pro-caspase-1. Inflammasomes activate pro-caspase-1 into its functional form, leading to the processing of pro-IL-1β and pro-IL-18 into the secreted forms IL-1β and IL-18, respectively. Active caspase-1 also starts the pathway to pyroptosis of infected cells (Bergsbaken et al. 2009).
Influenza A activates NLRP3 differently to the canonical and non-canonical pathways known to date. It causes the formation of the ZBP1-NLRP3 inflammasome to activate Z-DNA-binding protein 1 (ZBP1)-mediated pyroptosis and caspase-1 cleavage. Specifically, ZBP1 recruits RIPK3 and caspase-8 to assemble the ZBP1-NLRP3 inflammasome (Zheng et al. 2020). The influenza virus M2 proton channel (Ichinohe et al. 2010) and the PB1-F2 frameshift mutant (McAuley et al. 2013) have been associated with inflammasome activation. However, another publication reported that PB1-F2 inhibits RIG-I signaling and NLRP3 inflammasome activation (Yoshizumi et al. 2014).
cGAS
The cGAS cytosolic DNA sensor is a further defense against viral infection. It acts through the stimulator of interferon genes (STING), activating IRF3 and NF-κB resulting in the production of type I IFN and inflammatory cytokines (Motwani et al. 2019). Initially, it was the herpes simplex virus that was found to trigger the cGAS-STING pathway (Li et al. 2013). However, influenza A, though an RNA virus, also activates the pathway via the M2 protein that affects the translocation of mitochondrial DNA (mtDNA) into the cytosol (Moriyama et al. 2019).
Immune Cell Response to Influenza A
Airway epithelial cells are infected first by influenza viruses, triggering the secretion of antiviral and chemotactic molecules such as type I IFNs, IL-6, IL-8, TNF-α, CCL2 (MCP-1), RANTES, and MIP-1α that then recruit innate immune cells including natural killer (NK) cells, neutrophils, macrophages, and DCs (Veckman et al. 2006). After the initial innate immune system activation, the adaptive immune cells join the response to achieve final clearance of the influenza virus and establish immune memory to enhance the defense against future infections.
Innate Immune Cells
Natural Killer Cells, Neutrophils, and Macrophages
NK cells can eliminate influenza-infected cells by binding via NKp44 and NKp46 receptors which recognize HA (Mendelson et al. 2010) and proceed to lyse the target cells with granzyme and perforin. A unique lung resident CD56brightCD49a+CD103+CD69+ NK cell population was suggested to be responsible (Cooper et al. 2018). Alternatively, antibody-coated infected cells will be bound with CD16 by NK cells to induce antibody-dependent cellular cytotoxicity (ADCC) (Jegaskanda et al. 2019). Neutrophils, attracted by chemokines, migrate to the site of infection, and interact with the endothelium, leading to the initiation of phagocytosis, degranulation, and the deployment of neutrophil extracellular traps (NETs). The density of neutrophils in the lung correlates with the severity of influenza A infection (Camp and Jonsson. 2017). Additionally, neutrophils guide influenza-specific CD8? T cells to the airways via the chemokine CXCL12 (Lim et al. 2015). As the name implies, alveolar macrophages are situated in the lung where they contribute to lung homeostasis and switch to phagocytosing pathogens and infected cells during disease. They are also the main initial producers of type I IFNs in the lung during infection with RNA viruses (Kumagai et al. 2007). In influenza A virus infection, alveolar macrophages increase the resistance to infection of type 1 alveolar epithelial cells (Cardani et al. 2017).
Dendritic Cells
Dendritic cells, while considered to be part of the innate immune system, are a key connection to the adaptive immune system due to their role in priming and activation of naïve T cells. Depending on the state of health in the lung, up to three classes of DC are active: migratory conventional DCs (cDCs), and pDCs during normal conditions, with inflammatory stimuli generating monocyte-derived DCs (MoDCs) (Lin et al. 2008, Granot et al. 2017). Upon influenza infection, lung-based migratory cDCs take up viral antigens and migrate to the draining mediastinal lymph nodes (mLNs) to present antigens to naïve T cells. While both cDC1s and cDC2s can induce CD4+ and CD8+ T cell activation, cDC1s are considered to be more effective at presenting influenza antigens to CD8+ T cells (Ng et al. 2018). It has been suggested that this is due to the cDC1 subtype being more likely to be infected by the influenza A virus than cDC2s and pDCs. However, some disagreement remains on how the cDC1s acquired the antigen, via infection or by phagocytic uptake (Hao et al. 2008, Helft et al. 2012). The roles of pDCs are less clearly defined, as pDC-deficient mice showed delayed recruitment of T cells to the bronchoalveolar space but no difference in their influenza-specific CD8+ effector T cells (Wolf et al. 2009). MoDC proliferate during influenza infection and their presence contributes to infection resistance, but their specific function has not been clearly defined (Cruz et al. 2017).
Adaptive Immune Cells
CD8+ T Cells
CD8+ T cells are the key cell type of the adaptive immune system for clearing influenza virus infection. Naïve CD8+ T cells encounter peptides derived from viral proteins in the context of MHC class I molecules on the surface of antigen-presenting cells, such as DCs. This induces proliferation and differentiation of naïve CD8+ T cells into fully functional cytotoxic T lymphocytes (CTLs) (Ho et al. 2011, Kim and Braciale 2009). The expression of CCR7 (CD197) is downregulated while CXCR3 and CCR4 (CD194) expression is upregulated to facilitate migration from lymph nodes to the lungs to clear influenza A infected cells.
To achieve this, cytotoxic granules that contain perforin and granzymes, GrA and GrB, are released by CD8+ T cells. Perforin attacks infected target cells forming pores through which granzymes can diffuse to induce apoptosis. (Kreijtz et al. 2011); granzymes also have additional proteolytic functions (van Domselaar and Bovenschen 2011).
Further clearance mechanisms function via TNF receptor family-dependent pathways. CD8+ T cells express the Fas ligand (FasL, CD178, CD95L) that binds to Fas (CD95) on target cells inducing apoptosis via activation of caspases. TNF-related apoptosis-inducing ligand (TRAIL/CD253), found on CD8+ T cells, is another mechanism for CD8+ T cell-mediated apoptosis induction (Schmidt and Varga 2018).
CD4+ T Cells
Similarly, CD4+ T cells are also activated by DCs arriving from the lung into the draining lymph nodes during influenza A virus infection (Lukens et al. 2009). The cytokine environment created by DCs, epithelial cells, and inflammatory cells activates CD4+ T cells and drives them to a T helper 1 (Th1) phenotype. Then they secrete IL-2 and TNF-α in the draining lymph node and IFN-γ and IL-10 in the lungs, enhancing CD8+ T cells responses, activating macrophages, and triggering activation and differentiation of antibody-producing B cells to switch to IgG2a isotype antibodies (Brown et al. 2012, Baumgarth and Kelso 1996). Th2 type CD4+ T cells release IL-4, IL-5, and IL-13 and drive IgG1 and IgE antibody production in B cells (Brown et al. 2006). However, Th1 CD4+ T cells are more critical to recovery from influenza infection than the Th2 type. Finally, influenza infection also induces regulatory T CD4+ cells (Tregs), likely roles in modulating immunosuppression and tissue repair via IL-10 (Betts et al. 2012).
B Cells
Innate-like B cells (B-1 cells), in the bone marrow and spleen, secrete polyvalent IgM in the absence of infection that can detect pathogens including several influenza strains. This background IgM has neutralizing activity via complement recruitment (Choi et al. 2012). Driven by type I IFN response derived from innate immune cells, B-1 cells accumulate in the draining mediastinal lymph nodes after influenza infection where they differentiate into IgM-producing cells. Conventional B cells (B-2), primed by influenza A antigens, accumulate at the T to B cell border in the lymph node where they are stimulated by T helper cells to differentiate into short-lived antibody-secreting plasmablasts or germinal center-derived plasma cells. While the former will secrete antibodies for three to five days, the latter will diverge into long-lived antibody-secreting cells and circulating memory B cells (Lam and Baumgarth 2019).
Key antibodies produced are those specific for the HA and NA proteins of the influenza virus as these are involved in viral cell entry and subsequent release, respectively. Neutralization occurs via antibodies binding the HA globular head and blocking the attachment of the virus envelope to the host cell surface (Neu et al. 2016). In contrast, NA-specific antibodies are not neutralizing but instead block the enzymatic function of NA (Kreijtz et al. 2011). Other viral targets of the humoral immune response are antibodies to the M2 protein and even the virus's internal NP (Treanor et al. 1990, Carragher et al. 2008). Antibodies are also the triggering component in NK cell-mediated antibody-dependent cell cytotoxicity and Fc receptor-mediated innate immune cell phagocytosis (Jegaskanda et al. 2013).
Outlook
While it may seem that a lot is known about the biology of the influenza viruses and the multi-layered immune system response against them, influenza infections are still a major public health burden. A report in 2019 stated the estimated annual death toll from influenza-associated respiratory causes was 290,000-650,000, up from 250,000-500,000 in total (Iuliano et al. – Global Seasonal Influenza-associated Mortality Collaborator Network 2018). A separate study put the directly attributable death toll of influenza lower respiratory tract infections at 99,000-200,000 (GBD 2017 Influenza Collaborators 2019). While these data are not broken down by year, or influenza strain, they do underline the high burden on society, especially on the over 65s who represent more than 70% of cases.
Efforts to develop a universal vaccine to multiple subtypes to protect those most at risk is hampered by the virus’ ability to change its antigens. This ability to evolve and change antigens ensures influenza survival in the global population. Issues that need conquering are the immunogenicity of vaccines and their effectiveness at inducing cell-mediated immunity. Even then, the absence of the proofreading ability of the influenza virus’ RNA polymerase will always drive high mutation rates and push the evolution of the virus. It is likely that we will keep on requiring new therapeutics and vaccines, and immunology research will always be necessary to keep pace with the influenza virus subtype generation.
Discover an industry-leading portfolio of antibodies for flow cytometric analysis of the innate and adaptive immune system for a variety of species in our online catalog. Specifically, review our T cell, B cell, macrophage and dendritic cell, NK cell, and PRR antibodies. All validated antibodies with decades of peer-reviewed use in scientific journals are available in multiple formats to enable multicolor flow cytometry staining. Finally, also find a range of influenza A-specific antibodies.
References
- Baumgarth N and Kelso A. (1996). In vivo blockade of gamma interferon affects the influenza virus-induced humoral and the local cellular immune response in lung tissue. J Virol. 70, 4,411–4,418.
- Bergsbaken T et al. (2009). Pyroptosis: host cell death and inflammation. Nat Rev Microbiol. 7, 99–109.
- Betts RJ et al. (2012). Influenza A virus infection results in a robust, antigen-responsive, and widely disseminated Foxp3+ regulatory T cell response. J Virol. 86, 2,817–2,825.
- Bouvier NM and Palese P. (2008). The biology of influenza viruses. Vaccine 26 Suppl 4, D49–D53.
- Brown DM et al. (2006). CD4 T cell-mediated protection from lethal influenza: perforin and antibody-mediated mechanisms give a one-two punch. J Immunol. 177, 2,888–2,898.
- Brown DM et al. (2012). Multifunctional CD4 cells expressing gamma interferon and perforin mediate protection against lethal influenza virus infection. J Virol. 86, 6,792–6,803.
- Camp JV and Jonsson CB. (2017). A role for neutrophils in viral respiratory disease. Front Immunol. 8, 550.
- Cardani A et al. (2017). Alveolar macrophages prevent lethal influenza pneumonia by inhibiting infection of type-1 alveolar epithelial cells. PLoS Pathog. 13, e1006140.
- Carragher DM et al. (2008). A novel role for non-neutralizing antibodies against nucleoprotein in facilitating resistance to influenza virus. J Immunol. 181, 4,168–4,176.
- Chan CM et al. (2016). Hemagglutinin of influenza A virus binds specifically to cell surface nucleolin and plays a role in virus internalization. Virology 494, 78–88.
- Choi YS et al. (2012). B-1 cells in the bone marrow are a significant source of natural IgM. Eur J Immunol. 42, 120–129.
- Cohen M et al. (2013). Influenza A penetrates host mucus by cleaving sialic acids with neuraminidase. Virol J. 10, 321.
- Cooper GE et al. (2018). Human CD49a+ lung natural killer cell cytotoxicity in response to influenza A virus. Front Immunol. 9, 1,671.
- Cruz JL et al. (2017). Monocyte-derived dendritic cells enhance protection against secondary influenza challenge by controlling the switch in CD8+ T-cell immunodominance. Eur J Immunol. 47, 345–352.
- de Vries E et al. (2011). Dissection of the influenza A virus endocytic routes reveals macropinocytosis as an alternative entry pathway. PLoS Pathog. 7, e1001329.
- Dias A et al. (2009). The cap-snatching endonuclease of influenza virus polymerase resides in the PA subunit. Nature. 458, 914–918.
- Dumm RE and Heaton NS. (2019). The development and use of reporter influenza B viruses. Viruses. 11, 736.
- Ehre C et al. (2012). Overexpressing mouse model demonstrates the protective role of Muc5ac in the lungs. Proc Natl Acad Sci USA. 109, 16,528–16,533.
- Eierhoff T et al. (2010). The epidermal growth factor receptor (EGFR) promotes uptake of influenza A viruses (IAV) into host cells. PLoS Pathog. 6, e1001099.
- Fontana J et al. (2012). Structural changes in influenza virus at low pH characterized by cryo-electron tomography. J Virol. 86, 2,919–2,929.
- Fujioka Y et al. (2018). A sialylated voltage-dependent Ca(2+) channel binds hemagglutinin and mediates influenza A virus entry into mammalian cells. Cell Host Microbe 23, 809–818.e5.
- GBD 2017 Influenza Collaborators. (2019). Mortality, morbidity, and hospitalisations due to influenza lower respiratory tract infections, 2017: an analysis for the Global Burden of Disease Study 2017. Lancet Respir Med. 7, 69–89.
- Granot T et al. (2017). Dendritic cells display subset and tissue-specific maturation dynamics over human life. Immunity 46, 504–515.
- Hao X et al. (2008). Differential response of respiratory dendritic cell subsets to influenza virus infection. J Virol. 82, 4,908–4,919.
- Helft J et al. (2012). Cross-presenting CD103+ dendritic cells are protected from influenza virus infection. J Clin Invest. 122, 4,037–4,047.
- Ho AW et al. (2011). Lung CD103+ dendritic cells efficiently transport influenza virus to the lymph node and load viral antigen onto MHC class I for presentation to CD8 T cells. J Immunol. 187, 6,011–6,021.
- Ichinohe T et al. (2010). Influenza virus activates inflammasomes via its intracellular M2 ion channel. Nat Immunol. 11, 404–410.
- Iuliano AD et al. - Global Seasonal Influenza-associated Mortality Collaborator Network (2018). Estimates of global seasonal influenza-associated respiratory mortality: a modelling study. Lancet 391, 1,285–1,300.
- Jegaskanda S et al. (2013). Antibody-dependent cellular cytotoxicity is associated with control of pandemic H1N1 influenza virus infection of macaques. J Virol. 87, 5,512–5,522.
- Jegaskanda S et al. (2019). Influenza virus infection enhances antibody-mediated NK cell functions via type I interferon-dependent pathways. J Virol. 93, e02090-18.
- Kato H et al. (2005). Cell type-specific involvement of RIG-I in antiviral response. Immunity 23, 19–28.
- Kim TS and Braciale TJ. (2009). Respiratory dendritic cell subsets differ in their capacity to support the induction of virus-specific cytotoxic CD8+ T cell responses. PloS One 4, e4204.
- Kreijtz JH et al. (2011). Immune responses to influenza virus infection. Virus Res. 162, 19–30.
- Kumagai Y et al. (2007). Alveolar macrophages are the primary interferon-alpha producer in pulmonary infection with RNA viruses. Immunity 27, 240–252.
- Lam JH and Baumgarth N. (2019). The multifaceted B cell response to influenza virus. J Immunol. 202, 351–359.
- Le Goffic R et al. (2006). Detrimental contribution of the Toll-like receptor (TLR)3 to influenza A virus-induced acute pneumonia. PLoS Pathog. 2, e53.
- Le Goffic R et al. (2007). Cutting edge: Influenza A virus activates TLR3-dependent inflammatory and RIG-I-dependent antiviral responses in human lung epithelial cells. J Immunol. 178, 3,368–3,372.
- Li S et al. (2014). pH-controlled two-step uncoating of influenza virus. Biophys J. 106, 1,447–1,456.
- Li XD et al. (2013). Pivotal roles of cGAS-cGAMP signaling in antiviral defense and immune adjuvant effects. Science 341, 1,390–1,394.
- Lim K et al. (2015). Neutrophil trails guide influenza-specific CD8? T cells in the airways. Science 349, aaa4352.
- Lin KL et al. (2008). CCR2+ monocyte-derived dendritic cells and exudate macrophages produce influenza-induced pulmonary immune pathology and mortality. J Immunol. 180, 2,562–2,572.
- Londrigan SL et al. (2011). N-linked glycosylation facilitates sialic acid-independent attachment and entry of influenza A viruses into cells expressing DC-SIGN or L-SIGN. J Virol. 85, 2,990–3,000.
- Lukens MV et al. (2009). Respiratory syncytial virus-induced activation and migration of respiratory dendritic cells and subsequent antigen presentation in the lung-draining lymph node. J Virol. 83, 7,235–7,243.
- Mendelson M at al. (2010). NKp46 O-glycan sequences that are involved in the interaction with hemagglutinin type 1 of influenza virus. J Virol. 84, 3,789–3,797.
- McAuley JL et al. (2013). Activation of the NLRP3 inflammasome by IAV virulence protein PB1-F2 contributes to severe pathophysiology and disease. PLoS Pathog. 9, e1003392.
- McAuley JL et al. (2017). The cell surface mucin MUC1 limits the severity of influenza A virus infection. Mucosal Immunol. 10, 1,581–1,593.
- Moriyama M et al. (2019). Influenza A virus M2 protein triggers mitochondrial DNA-mediated antiviral immune responses. Nat Commun. 10, 4,624.
- Motwani M et al. (2019). DNA sensing by the cGAS-STING pathway in health and disease. Nat Rev Genet. 20, 657–674.
- Neu KE et al. (2016). Heads, stalks and everything else: how can antibodies eradicate influenza as a human disease? Curr Opin Immunol. 42, 48–55.
- Ng SL et al. (2018). Type 1 conventional CD103+ dendritic cells control effector CD8+ T cell migration, survival, and memory responses during influenza infection. Front Immunol. 9, 3,043.
- Ng WC et al. (2014). The macrophage galactose-type lectin can function as an attachment and entry receptor for influenza virus. J Virol. 88, 1,659–1,672.
- Nicholls JM et al. (2007). Tropism of avian influenza A (H5N1) in the upper and lower respiratory tract. Nat Med. 13, 147–149.
- Onomoto K et al. (2012). Critical role of an antiviral stress granule containing RIG-I and PKR in viral detection and innate immunity. PloS One 7, e43031.
- Pflug A et al. (2017). Structural insights into RNA synthesis by the influenza virus transcription-replication machine. Virus Res. 234, 103–117.
- Pothlichet J et al. (2013). Type I IFN triggers RIG-I/TLR3/NLRP3-dependent inflammasome activation in influenza A virus infected cells. PLoS Pathog. 9, e1003256.
- Poux C et al. (2019). A single-stranded oligonucleotide inhibits Toll-like receptor 3 activation and reduces influenza A (H1N1) infection. Front. Immunol. 10, 2,161.
- Roy AM et al. (2000). Early stages of influenza virus entry into Mv-1 lung cells: involvement of dynamin. Virology 267, 17–28.
- Rust MJ et al. (2004). Assembly of endocytic machinery around individual influenza viruses during viral entry. Nat Struct Mol Biol. 11, 567–573.
- Samji T. (2009). Influenza A: understanding the viral life cycle. Yale J Biol Med. 82, 153–159.
- Schmidt ME and Varga SM (2018). The CD8 T cell response to respiratory virus infections. Front. Immunol. 9, 678.
- Schoggins JW et al. (2014). Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature 505, 691–695.
- Sims LD et al. (2005). Origin and evolution of highly pathogenic H5N1 avian influenza in Asia. Vet Rec. 157, 159–164.
- Sun L et al. (2013). Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339, 786–791.
- Treanor JJ et al. (1990). Passively transferred monoclonal antibody to the M2 protein inhibits influenza A virus replication in mice. J Virol. 64, 1,375–1,377.
- Trombetta CM et al. (2019). Influenza D virus: serological evidence in the Italian population from 2005 to 2017. Viruses 12, 30.
- Upham JP et al. (2010). Macrophage receptors for influenza A virus: role of the macrophage galactose-type lectin and mannose receptor in viral entry. J Virol. 84, 3,730–3,737.
- van Domselaar R and Bovenschen N (2011). Cell death-independent functions of granzymes: hit viruses where it hurts. Rev Med Virol. 21, 301–314.
- van Riel D et al. (2006). H5N1 virus attachment to lower respiratory tract. Science 312, 399.
- van Riel D et al. (2007). Human and avian influenza viruses target different cells in the lower respiratory tract of humans and other mammals. Am J Pathol. 171, 1,215–1,223.
- van Riel D et al. (2010). Seasonal and pandemic human influenza viruses attach better to human upper respiratory tract epithelium than avian influenza viruses. Am J Pathol. 176, 1,614–1,618.
- Veckman V et al. (2006). TNF-alpha and IFN-alpha enhance influenza-A-virus-induced chemokine gene expression in human A549 lung epithelial cells. Virology 345, 96–104.
- Vijaykrishna D et al. (2010). Reassortment of pandemic H1N1/2009 influenza A virus in swine. Science 328, 1,529.
- Webb LG and Fernandez-Sesma A. (2022). RNA viruses and the cGAS-STING pathway: reframing our understanding of innate immune sensing. Curr Opin Virol. 53, 101206.
- Wolf AI et al. (2009). Plasmacytoid dendritic cells are dispensable during primary influenza virus infection. J Immunol. 182, 871–879.
- Yoshizumi T et al. (2014). Influenza A virus protein PB1-F2 translocates into mitochondria via Tom40 channels and impairs innate immunity. Nat Commun. 5, 4,713.
- Zheng M et al. (2020). Caspase-6 is a key regulator of innate immunity, inflammasome activation, and host defense. Cell 181, 674–687.e13.






