Dengue Virus
What Is Dengue Virus
The dengue virus (DENV), first isolated in 1943, is assigned to the family Flaviviridae, genus Flavivirus. It has four serotypes (DENV-1, DENV-2, DENV-3, and DENV-4) and transmission to humans occurs via the Aedes mosquito, mainly Aedes aegypti. Its global reach has mirrored the spread of Aedes aegypti in tropical and subtropical areas (Messina et al. 2014). It is suggested that 390 million dengue infections occur per year, of which 96 million develop symptoms (Bhatt et al. 2013).
Dengue Fever
After being exposed to the virus (of any DENV serotype) through a mosquito bite, an incubation period of 4-7 days follows. Then symptoms tend to be noticed for the next 5 days, but not all infected individuals show signs of illness (Bhatt et al. 2013). This is called dengue fever (DF) and is similar to mild flu-like syndrome. Some hemorrhagic manifestations, such as petechiae, may occur. The next form is dengue hemorrhagic fever (DHF) which adds thrombocytopenia, coagulopathy, and vascular fragility and permeability. The most severe form, dengue shock syndrome (DSS), is characterized by a rapid, weak pulse, and may progress to hypovolemic shock and death (Carlos et al. 2005).
Dengue Virion
DENV has an outer icosahedral shell and a lipid bilayer that surrounds the nucleocapsid which contains the 11,000 kb positive- and single-stranded RNA (ssRNA) genome with a single open reading frame (Kuhn et al. 2002) that codes for:
-
Three structural proteins
- C – capsid
- E – envelope
- prM, further processed to M – membrane
-
Seven nonstructural proteins
- NS1, NS2a, NS2b, NS3, NS4a, NS4b, and NS5
The E protein forms the outer layer (90 E fusion protein pairs) and also binds the cellular receptors and initiates fusion with the host cell (Chambers et al. 1990). The next lower layer is made up of the M protein. In the immature DENV, the prM protein forms a trimer with the E protein pairs, resulting in the appearance of spikes on the virion surface (Zhang et al. 2003). At the core the C protein forms the container for the viral RNA.
The NS proteins have roles in viral replication and host immune system evasion. Of these, NS3 and NS5 are better defined than the other NS proteins. NS3 is a viral protease and helicase, which cleaves the single viral polypeptide into the seven proteins and unwinds the RNA duplex during replication, respectively. NS2b serves as a cofactor for the protease activity (Falgout et al. 1991) while NS4b interacts with the helicase domain (Umareddy et al. 2006). NS5 is the RNA-dependent RNA polymerase that also functions as a methyltransferase, which caps the 5′-RNA of new viral genomes (Issur et al. 2009). The functions of the NS1 and the transmembrane proteins NS2a, NS2b, NS4a, and NS4b are less well defined. NS1 is essential for viral replication (Lindenbach and Rice 1999) and together with NS4a and NS4b, aids in vesicle formation for the viral replication complex (RC) (Watterson et al. 2016). NS2a is required for viral RNA synthesis and virion assembly (Xie et al. 2015).
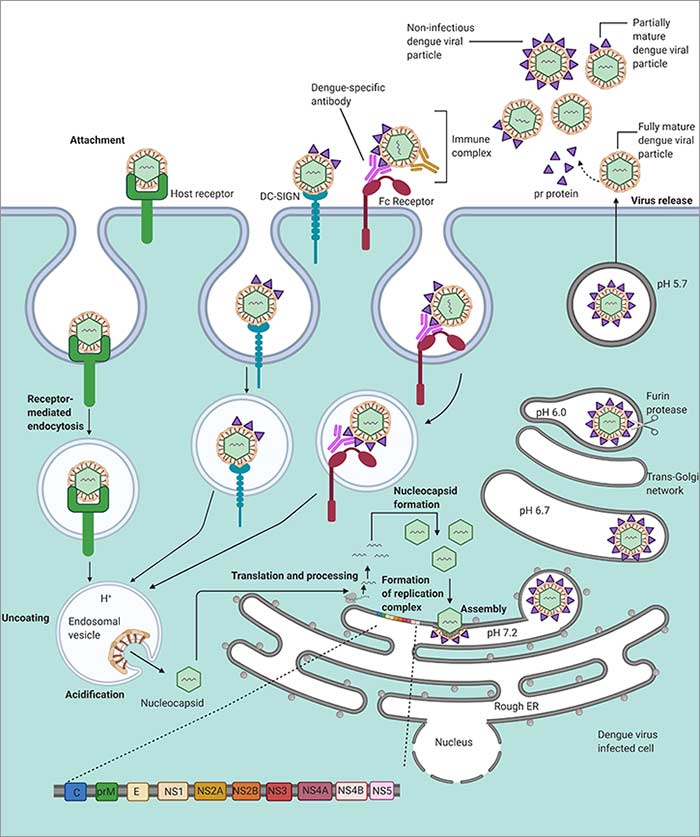
Dengue virus life cycle.
DENV Lifecycle
The Aedes mosquito is the vector that spreads DENV to humans. The precise binding receptors have not been fully defined yet, but candidate receptors are: heparan sulfate, glycosphingolipid nLc4Cer, DC-SIGN, mannose, CD14, and HSP70/90 (Cruz-Oliveira et al. 2015). Similarly, several cell types have been found to be permissive for DENV infection; they are: dendritic cells (DCs), endothelial cells, fibroblasts, keratinocytes, macrophage, mast cells, and monocytes (Garcia et al. 2017). The replication steps have been defined as:
- E protein initiates cell entry via clathrin mediated endocytosis
- Viral genome is released into the cytoplasm
- The RC forms and the unraveled viral RNA is translated by the ribosomes
- Viral and host proteases process the viral polypeptide chain
- prM and E proteins assemble in the endoplasmic reticulum
- Viral RNA is synthesized
- Viral assembly proceeds in the Golgi apparatus
- prM is processed to the mature form
- Mature and some immature viruses are shed from the host cell
(Mackenzie 2005, Mukhopadhyay et al. 2005, Welsch et al. 2009)
Immune Response
On cell entry, the DENV RNA will be detected in innate immune cells by the pattern recognition receptors (PRR), retinoic acid-inducible gene I (RIG-I), melanoma differentiation-associated protein 5 (MDA5), Toll-like receptor 3 (TLR3), and TLR7 (Nasirudeen et al. 2011, Wang et al. 2006). Activation of these receptors will trigger type 1 interferon (IFN) responses. The mannose-binding lectin (MBL) recognizes mannose glycans on DENV and activates the classical complement cascade resulting in the formation of the membrane attack complex (MAC) to lyse virions, recruit phagocytes, and drive inflammation (Fujita et al. 2004).
Antibodies and Antigens
Our recombinant DENV NS1 proteins are available in their native folded state complete with post-translational modifications. This allows them to deliver optimal antigenicity and makes them suitable for use in vaccine research and serology-based assays. The recombinant DENV NS1 proteins are available in all four serotypes.
Bio-Rad also offers a DENV type 2 inactivated pathogen along with two antibodies to the virus. 2645-5057, clone 3H5.1, in purified format, works in immunofluorescence and recognizes the dengue virus type 2. MCA2277, clone dengue 1-11 (3), also available in purified format, binds all four serotypes of DENV and can be used in ELISA, immunofluorescence, western blotting, and in immunohistology on paraffin embedded tissue sections. Additionally, an antibody, clone 6b6c-1, to the Saint Louis Encephalitis virus strain (MSI-7) envelope glycoprotein is available. This clone also reacts with other members of Flaviviridae including; Dengue 1 (Hawaii), Dengue 2 (New Guinea C), Dengue 3 (H87), and Dengue 4 (H241).
Four Serotypes of Dengue Virus (DENV) Non-Structural Protein 1 (NS1)
| Product Code | Specificity | Format | Target | Applications |
|---|---|---|---|---|
| PIP047B | DENGUE Virus Type 1 NS1 Antigen | Rec. Protein | Viral | E |
| PIP048B | DENGUE Virus Type 2 NS1 Antigen | Rec. Protein | Viral | E |
| PIP049A | DENGUE Virus Type 3 NS1 Antigen | Rec. Protein | Viral | E |
| PIP050B | DENGUE Virus Type 4 NS1 Antigen | Rec. Protein | Viral | E |
Additional Dengue Virus Related Products
Inactivated Pathogen
| Product Code | Specificity | Format | Target | Applications |
|---|---|---|---|---|
| PIP006 | DENGUE Virus Type 2 antibody | Inactivated Pathogen | Viral | E |
Antibodies to Dengue Virus
| Product Code | Specificity | Target | Host | Isotype | Clone | Applications |
|---|---|---|---|---|---|---|
| 2645-5057 | DENGUE Virus antibody: Purified | Viral | Mouse | IgG1 | 3H5.1 | IF |
| MCA2277 | DENGUE Virus antibody: Purified | Viral | Mouse | IgG2a | Dengue 1-11(3) | E, P*, WB |
| OBT1676 | ST. Louis Encephalitis Virus antibody: Purified | Viral | Mouse | IgG2a | 6b6c-1 | E, IF |
References
- Bhatt S et al. (2013). The global distribution and burden of dengue. Nature. 496(7446), 504-507.
- Carlos CC et al. (2005). Comparison of clinical features and hematologic abnormalities between dengue fever and dengue hemorrhagic fever among children in the Philippines. Am J Trop Med Hyg. 73(2), 435-440.
- Chambers TJ et al. (1990). Flavivirus genome organization, expression, and replication. Annu Rev Microbiol. 44, 649-688.
- Cruz-Oliveira C et al. (2015). Receptors and routes of dengue virus entry into the host cells. FEMS Microbiol Rev. 39(2), 155-170.
- Falgout B et al. (1991). Both nonstructural proteins NS2B and NS3 are required for the proteolytic processing of dengue virus nonstructural proteins. J Virol. 65(5), 2467-2475.
- Fujita T et al. (2004). The lectin-complement pathway--its role in innate immunity and evolution. Immunol Rev. 198, 185-202.
- Garcia M et al. (2017). Skin innate immune response to flaviviral infection. Eur Cytokine Netw. 28(2), 41-51.
- Issur M et al. (2009). The flavivirus NS5 protein is a true RNA guanylyltransferase that catalyzes a two-step reaction to form the RNA cap structure. RNA. 15(12), 2340-2350.
- Kuhn RJ et al. (2002). Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell. 108(5), 717-725.
- Lindenbach BD and Rice CM (1999). Genetic interaction of flavivirus nonstructural proteins NS1 and NS4A as a determinant of replicase function. J Virol. 73(6), 4611-4621.
- Mackenzie J (2005). Wrapping things up about virus RNA replication. Traffic. 6(11), 967-977.
- Messina JP et al. (2014). Global spread of dengue virus types: mapping the 70 year history. Trends Microbiol. 22(3), 138-146.
- Mukhopadhyay S et al. (2005). A structural perspective of the flavivirus life cycle. Nat Rev Microbiol. 3(1), 13-22.
- Nasirudeen AM et al. (2011). RIG-I, MDA5 and TLR3 synergistically play an important role in restriction of dengue virus infection. PLoS Negl Trop Dis. 5(1), e926.
- Umareddy I et al. (2006). Dengue virus NS4B interacts with NS3 and dissociates it from single-stranded RNA. J Gen Virol. 87(Pt 9), 2605-2614.
- Wang JP et al. (2006). Flavivirus activation of plasmacytoid dendritic cells delineates key elements of TLR7 signaling beyond endosomal recognition. J Immunol. 177(10), 7114-7121.
- Watterson D et al. (2016). The many faces of the flavivirus NS1 protein offer a multitude of options for inhibitor design. Antiviral Res. 130, 7-18.
- Welsch S et al. (2009). Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe. 5(4), 365-375.
- Xie X et al. (2015). Two distinct sets of NS2A molecules are responsible for dengue virus RNA synthesis and virion assembly. J Virol. 89(2), 1298-1313.
- Zhang Y et al. (2003). Structures of immature flavivirus particles. The EMBO journal. 22(11), 2604–2613.



