SARS-CoV-2 — the Deadly Coronavirus

- On This Page
- Coronaviruses
- What is SARS-CoV-2
- Spike protein structure
- Spike protein function
- Lifecycle
- Antibodies
- References
s
Introduction to ELISA Basics Guide

The ELISA Basics Guide has the right amount of detail to help you plan your experiment and achieve a successful ELISA.
s
About Coronaviruses
Coronaviruses, a large virus family with hundreds of members, are generally responsible for mild upper-respiratory tract illnesses, like the common cold. However, since the turn of the century, new coronaviruses have jumped species and led to serious morbidity and increased mortality.
The SARS coronavirus (SARS-CoV) caused severe acute respiratory syndrome (SARS) in 2002; yet disappeared by 2004. The MERS coronavirus (MERS-CoV) led to Middle East respiratory syndrome (MERS) in 2012 and has not been eliminated yet. Coronavirus disease 2019 (COVID-19), caused by SARS-CoV-2, broke out in 2019 and has led to a worldwide pandemic.
Coronavirus Components
Coronaviruses are positive-sense, single-stranded RNA viruses made up of four structural proteins:
- S (spike)
- E (envelope)
- M (membrane)
- N (nucleocapsid)
The S, E, and M proteins form the viral envelope, while the N protein stores the RNA genome. The S protein is the key protein mediating host cell infection. Additionally, up to 16 nonstructural proteins are synthesized to support regulatory and enzymatic functions.
What Is SARS-CoV-2?
In 2019, an acute respiratory disease began to spread in Wuhan, China. It was subsequently found to be caused by a coronavirus leading to the name — coronavirus disease 19 (COVID-19). It was the third time a coronavirus jumped species to infect humans, the previous outbreaks were SARS in 2002 and MERS in 2012. However, unlike SARS and MERS, COVID-19 has led to a pandemic lasting into 2021 (Zhou et al. 2020).
Coronaviruses (CoVs), part of the Coronaviridae family, are enveloped, positive-sense single-stranded RNA viruses. SARS-CoV-2, along with the other pathogenic CoVs belongs to the Betacoronavirus genus, group 2 (Coronaviridae Study Group of the International Committee on Taxonomy of Viruses 2020). SARS-CoV-2 shares ~79% sequence identity with SARS-CoV and ~50% with MERS-CoV.
14 open reading frames (ORFs) and 16 nonstructural proteins (nsp 1-16) that generate the replicase complex, take up the majority of the virus’s genome. The four structural proteins: spike (S), envelope (E), membrane (M), and nucleocapsid (N) and nine accessory proteins are encoded in the remaining third of the genome. The S protein mediates the virus cell entry during infection (Perlman and Netland 2009, Lu et al. 2020) and is the key focus of therapeutic and vaccine efforts.
Spike Protein Structure and Relevance to Host Cell Fusion
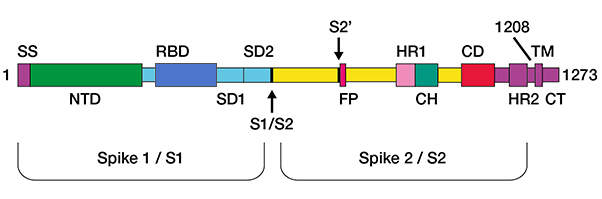
The spike protein (180–200 kD) (Figure 1), split into S1 and S2 sections, consists of a short intracellular C-terminal domain, a transmembrane (TM) segment in the viral membrane, and an extracellular N-terminus.
Fig. 1. Structure of the spike protein. SS, signal sequence; S2′, S2′ protease cleavage site; FP, fusion peptide; HR1, heptad repeat 1; CH, central helix; CD, connector domain; HR2, heptad repeat 2; TM, transmembrane domain; CT, cytoplasmic tail. Arrows indicate protease cleavage sites.
Modified from: Wrapp D et al. (2020).
The 1,273 amino acid (aa) long SARS-CoV-2 S protein has a signal peptide at the N-terminus (amino acids 1-13), the S1 subunit, needed for receptor binding, stretching from residue 14 to 685, and the S2 subunit involved in membrane fusion occupying aa 686 to1,273. The S subunits have further subdivisions (Xia S. et al. 2020).
-
S1 subunit:
- N-terminal domain (aa 14-305)
- Receptor-binding domain (RBD, aa 319-541)
-
S2 subunit:
- Fusion peptide (FP) (aa 788-806)
- Heptapeptide repeat sequence 1 (HR1) (aa 912-984)
- HR2 (aa 1163-1213)
- Transmembrane (TM) domain (aa 1,213-1,237)
- Cytoplasm domain (CT) (aa 1,237-1,273)
The RBD region of the S1 subunit (Figure 1) binds the host cell receptor, angiotensin-converting enzyme 2 (ACE2). Specifically, the receptor-binding motif, a portion of RBD, makes direct contacts with ACE2. The SARS-CoV-2 RBD has more residues that directly bind ACE2 than the original SARS-RBD (21 versus 17); meaning a larger surface interacts with ACE2 (Wrapp et al. 2020).
In the S2 subunit, the mainly hydrophobic FP sequence anchors SARS-CoV-2 to the host cell membrane, when the S protein changes from the stable prefusion to the intermediate conformation. HR1 and HR2 are made up of a repetitive heptapeptide and form a six-helical bundle (6-HB) with HRs from the other S proteins that make up the trimer spike on SARS-Cov-2. The 6-HB is also involved in viral fusion and entry of the S2 subunit into the host cell (Xia et al. 2020). HR1 is located near the hydrophobic FP at the C-terminus and HR2 is placed next to the TM domain at the N-terminus of S2. HR1 forms a homotrimeric structure exposing three highly conserved hydrophobic grooves that will bind to HR2. The TM domain anchors the S protein in the viral membrane and is followed by the CT domain (Robson 2020).
Spike Protein Function
After the S1 subunit has bound the ACE2 receptor, the prefusion S trimer destabilizes, leading to the cleavage of the S1 subunit at the S1/S2 boundary (Figure 1) (Walls et al. 2017) by furin (Johnson et al. 2021). This is followed by a subsequent cleavage at the S2’ site by the transmembrane serine protease 2 (TMPRSS2) (Bestle et al. 2020, Hoffmann et al. 2020a). The latter removes the N-terminal region of S2, freeing the FP to initiate the membrane fusion process. The FP achieves that by interacting with and rearranging the lipid bilayers of the host cell membrane (Millet and Whittaker 2018). The motion of the FP is effected by the HR1 domain inducing the unstable intermediate configuration of the S2 subunit, characterized by the exposed coiled-coil of the HR1 domain that induces interaction of the HR2 domain with the HR1 trimer to form 6-HB. This interaction pulls the viral envelope and cell membrane close together, initiating viral fusion, entry, and infection (Xia et al. 2018). The alternative, although minor (Shirato et al. 2018), infection path is via endosomes where endolysosomal cathepsin L activates the SARS-CoV-2 Spike (Hoffmann et al. 2020b).
SARS-CoV-2 Lifecycle
After fusion, the RNA genome is released into the cytosol, ORF1a and ORF1b are translated into viral replicase proteins (pp1a and pp1b). These are then processed into replicase complex nonstructural proteins (nsps). Among them the RNA-dependent RNA polymerase (RdRp) will synthesize new genomic RNA. The host cell endoplasmic reticulum (ER) buds off into virus-induced double-membrane vesicles (DMVs) where viral replication commences. The DMVs form larger aggregates that combine with newly synthesized positive-strand RNA to function as a template for full-length negative-strand RNA and subgenomic (sg)RNA. The translation of sgRNA generates the proteins, accessory and structural (i.e. N, S, M, and E), that are secreted into the ER–Golgi intermediate compartment (ERGIC) in preparation for virion assembly. Finally, positive-sense RNA is combined with the newly synthesized virions, which are released from the plasma membrane (Fehr and Perlman 2015).
SARS-CoV-2 Research Antibodies
Bio-Rad offers a range of rabbit polyclonal antibodies for SARS-CoV-2 research. The range is focused on the spike protein, with all antibodies suitable for use in ELISA, immunofluorescence, and western blotting. The antibodies enable detection of the SARS-CoV-2 spike protein subdomains: S1, S1 cleavage site, S1 RBD, S2 carboxy terminus, and S2 center section.
To complement that range, we also offer a mouse monoclonal ELISA sandwich antibody pair for measuring the nucleoprotein. Additionally, the detection antibody is suitable for immunofluorescence, and the capture antibody will detect nucleoprotein in western blots. Finally, a rabbit polyclonal antibody to the envelope protein for use in ELISA, immunofluorescence, and immunohistochemistry on paraffin embedded tissue sections is also available. See the table below for full ordering details.
Researching the infections caused by alphacoronaviruses and betacoronaviruses in felines, canines, and cattle? Read the micro-review on coronaviruses in veterinary species.
SARS-CoV-2 / COVID-19 Antibody Research Solutions
 Explore the varied solutions Bio-Rad provides to advance COVID-19 research, diagnostic, and vaccine development work. Ranging from a comprehensive immunology, adaptive and innate, antibody range, via a market beating secondary antibody portfolio, to a state-of-the-art custom antibody generation service Bio-Rad can help to support your scientific endeavours.
Explore the varied solutions Bio-Rad provides to advance COVID-19 research, diagnostic, and vaccine development work. Ranging from a comprehensive immunology, adaptive and innate, antibody range, via a market beating secondary antibody portfolio, to a state-of-the-art custom antibody generation service Bio-Rad can help to support your scientific endeavours.
SARS-CoV-2 Research Antibodies
| Description | Target | Format | Clone | Applications | Citations | Code |
|---|
References
- Bestle D et al. (2020). TMPRSS2 and furin are both essential for proteolytic activation of SARS-CoV-2 in human airway cells. Life Sci Alliance 3(9), e202000786.
- Coronaviridae Study Group of the International Committee on Taxonomy of Viruses (2020). The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nature Microbiol 5(4), 536-544.
- Fehr AR and Perlman S (2015). Coronaviruses: an overview of their replication and pathogenesis. Methods Mol Biol 1,282, 1-23.
- Hoffmann M et al. (2020a). A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol Cell 78(4), 779-784.e5.
- Hoffmann M et al. (2020b). SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181(2), 271-280.e8.
- Johnson BA et al. (2021). Loss of furin cleavage site attenuates SARS-CoV-2 pathogenesis. Nature 10.1,038/s41586-021-03237-4.
- Lu R et al. (2020). Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 395(10,224), 565-574.
- Millet JK and Whittaker GR (2018). Physiological and molecular triggers for SARS-CoV membrane fusion and entry into host cells. Virology 517, 3-8.
- Perlman S and Netland J (2009). Coronaviruses post-SARS: update on replication and pathogenesis. Nat Rev Microbiol 7(6), 439-450.
- Robson B (2020). Computers and viral diseases. Preliminary bioinformatics studies on the design of a synthetic vaccine and a preventative peptidomimetic antagonist against the SARS-CoV-2 (2019-nCoV, COVID-19) coronavirus. Comput Biol Med 119, 103,670.
- Shirato K et al. (2018). Wild-type human coronaviruses prefer cell-surface TMPRSS2 to endosomal cathepsins for cell entry. Virology 517, 9-15.
- Walls AC et al. (2017). Tectonic conformational changes of a coronavirus spike glycoprotein promote membrane fusion. Proc Natl Acad Sci USA 114(42), 11,157-11,162.
- Wrapp D et al. (2020). Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 367(6,483), 1,260-1,263.
- Xia S et al. (2020). Fusion mechanism of 2019-nCoV and fusion inhibitors targeting HR1 domain in spike protein. Cellular Mol Immunol 17(7), 765-767.
- Xia S et al. (2018). Peptide-based membrane fusion inhibitors targeting HCoV-229E spike protein HR1 and HR2 domains. Int J Mol Sci 19(2), 487.
- Zhou P et al. (2020). A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579(7,798), 270-273.