C9orf72 in Amyotrophic Lateral Sclerosis and Frontotemporal Dementia
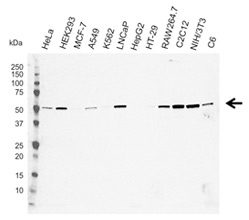
Fig. 1. Western blot analysis of whole cell lysates probed with C9orf72 antibody followed by detection with HRP conjugated Goat Anti-Mouse IgG (1/5,000, STAR207P) and visualized on the ChemiDoc MP Imaging System with 73 sec exposure. Arrow points to C9orf72 (molecular weight 54 kDa).
Amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD) are devastating and fatal neurodegenerative diseases with similarities in pathology, genetics, and neurobehavioral aspects. FTD manifests itself as a degeneration of the frontal and temporal lobe regions, leading to cognitive decline and behavioral impairment. ALS is a motor neuron disease characterized by degeneration in upper and lower motor neurons (Rowland and Shneider 2001). Onset of either can range from 27 to 83 years of age with mortality generally occurring 2-5 years after onset, although the disease can last up to twenty years (Majounie et al. 2012, Woollacott and Mead 2014).
Link to C9orf72
A key event in ALS and FTD research was the discovery of the C9orf72 gene as the most frequent genetic association with the diseases. In particular, a hexanucleotide GGGGCC (G4C2) repeat expansion in the C9orf72 gene is the trigger (DeJesus-Hernandez et al. 2011, Renton et al. 2011). This discovery applies mainly to Europeans and Americans, as it is rarely seen in Asia. The G4C2 repeat expansion of C9orf72 accounts for approximately 40% of hereditary ALS and 18% of familial FTD cases (Renton et al. 2014). C9orf72 is widely expressed and can be detected as a 54 kDa band in western blots (Figure 1).
Relevance of G4C2 Repeats
To date it has not been possible to define a threshold for pathological repeat-length. Most healthy individuals tend to have less than 11 repeats. However, disease may occur after a provisional threshold of 30 repeats. In ALS patients, repeat numbers have been found to range from 700 to 1600 (Rutherford et al. 2012). Further variability is caused by the instability of the hexanucleotide repeats, while the central nervous system may have very large numbers of repeats, blood samples can show far lower numbers. Cases have been found where a 70-repeat segment in a parent did not cause ALS or FTD but caused a larger expansion, and ALS, in the next generation (Xi et al. 2015).
The G4C2 repeats can be transcribed bidirectionally into repetitive RNA, forming sense and antisense RNA foci (Mizielinska et al. 2013). It has been shown that both RNA species are translated to generate five distinct dipeptide repeat proteins (DPRs) using a non-canonical mechanism classified as repeat-associated non-ATG (RAN) translation (Ash et al. 2013, Mori et al. 2013).
The pathology of the C9orf72 gene mutation is still being investigated based on three suggested scenarios:
- Loss of function of C9orf72 protein
- Toxic gain of function from sense and antisense C9orf72 repeat RNA
- Effects of DPRs
These are not expected to be mutually exclusive combinations.
Effects on Immune Cells
Studies in mice showed that C9orf72 expression is high in myeloid cells, and in the brain the highest levels are seen in microglia, suggesting a role for immune homeostasis. Knockout studies found elevated levels of the autophagy related proteins p62 and LC3. In spleen, the macrophage/monocyte marker Trem2 showed increased expression, so did the inflammatory cytokines IL-1 beta, IL-6, and IL-10. Additionally, immune responses in macrophages and microglia were altered leading to age related neuroinflammation typical of C9orf72 ALS (O'Rourke et al. 2016), fitting into the loss of function proposal (Lall and Baloh 2017).
Toxic Gain of Function in ALS/FTD
A further scenario envisions that the translation of G4C2 repeats in the promotor region results in a downregulation of C9orf72, thus abrogating its function. Experimental data has shown that ALS sufferers have decreased C9orf72 transcript levels (DeJesus-Hernandez et al. 2011). Work in model animals supports this hypothesis (Ciura et al. 2013, Therrien et al. 2013). Additional RNA-mediated toxicity has been put forward on the basis of observed positive and negative strand RNA foci in the frontal cortex, spinal cord, and cerebellum (DeJesus-Hernandez et al. 2011, Gendron et al. 2013). These RNAs would dock with RNA-binding proteins and interfere in the cell’s RNA metabolism. A final toxic gain of function has been ascribed to DPRs which accumulate in the brains of ALS patients, and could then drive neurodegeneration. It has been found that DPR accumulation and RNA inclusion can damage nucleocytoplasmic transport and lead to neural death (Jovicic et al. 2015, Zhang et al. 2015).
DPRs, TDP-43, and C9orf72 Mutation
Neurons and glia in most ALS patients and approximately half of the FTD cases show inclusions of the RNA-binding protein TDP-43 (TAR DNA-binding protein 43), highlighting a possible shared pathway of ALS and FTD (Ling et al. 2013, Mackenzie et al. 2013). The inclusions can be found in several areas of the brain. Appearance of TDP-43 inclusions is suggested to represent the damage caused by DPRs as it occurs after DPR formation and accumulation (Mackenzie et al. 2013).
Despite the high level of translational research that has been brought to bear on ALS and FTD, no reliable diagnostic tools are available, neither are effective therapeutics on the horizon for clinical use. Further applied research is needed to profile the effects of G4C2 repeats in the C9orf72 mutation and other candidate genes.
Antibodies to C9orf72 and TDP-43
Bio-Rad, a leading supplier of western blotting equipment and reagents, has antibodies available for detecting C9orf72 and TDP-43. These reagents are from the Precision Ab Antibody portfolio and are validated against multiple standard cell lysates in western blotting.
References
- Ash PE et al. (2013). Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron 77, 639–646.
- Ciura S et al. (2013). Loss of function of C9orf72 causes motor deficits in a zebrafish model of amyotrophic lateral sclerosis. Ann Neurol. 74(2), 180-187.
- DeJesus-Hernandez M et al. (2011). Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 72(2), 245-256.
- Gendron TF et al. (2013). Antisense transcripts of the expanded C9ORF72 hexanucleotide repeat form nuclear RNA foci and undergo repeat-associated non-ATG translation in c9FTD/ALS. Acta Neuropathol. 126(6), 829-844.
- Jovicic A et al. (2015). Modifiers of C9orf72 dipeptide repeat toxicity connect nucleocytoplasmic transport defects to FTD/ALS. Nat Neurosci. 18(9), 1226-1229.
- Lall D and Baloh RH (2017). Microglia and C9orf72 in neuroinflammation and ALS and frontotemporal dementia. J Clin Invest. 127(9), 3250-3258.
- Ling SC et al. (2013). Converging mechanisms in ALS and FTD: disrupted RNA and protein homeostasis. Neuron 79(3), 416-438.
- Mackenzie IR et al. (2013). Dipeptide repeat protein pathology in C9ORF72 mutation cases: clinico-pathological correlations. Acta Neuropathol. 126(6):859-879.
- Majounie E et al. (2012). Frequency of the C9orf72 hexanucleotide repeat expansion in patients with amyotrophic lateral sclerosis and frontotemporal dementia: a cross-sectional study. Lancet Neurol. 11(4):323-330.
- Mizielinska S et al. (2013). C9orf72 frontotemporal lobar degeneration is characterised by frequent neuronal sense and antisense RNA foci. Acta Neuropathol. 126(6), 845-857.
- Mori K et al. (2013). The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science 339, 1335–1338.
- Neumann M et al. (2006). Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 314(5796), 130-133.
- O'Rourke JG et al. (2016). C9orf72 is required for proper macrophage and microglial function in mice. Science 351(6279), 1324-1329.
- Renton AE et al. (2011). A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS–FTD. Neuron 72:257–268.
- Renton AE et al. (2014). State of play in amyotrophic lateral sclerosis genetics. Nat Neurosci. 17(1), 17-23.
- Rowland LP and Shneider NA (2001). Amyotrophic lateral sclerosis. New Engl. J. Med. 344, 1688–1700.
- Rutherford NJ et al. (2012). Length of normal alleles of C9ORF72 GGGGCC repeat do not influence disease phenotype. Neurobiol. Aging 33, 2950.e5–7.
- Therrien M et al. (2013). Deletion of C9ORF72 results in motor neuron degeneration and stress sensitivity in C. elegans. PLoS One 8(12):e83450.
- Woollacott IO and Mead S (2014). The C9ORF72 expansion mutation: gene structure, phenotypic and diagnostic issues. Acta Neuropathol. 127(3):319-332.
- Xi Z et al. (2015). Jump from pre-mutation to pathologic expansion in C9orf72. Am J Hum Genet. 96(6), 962-970.
- Zhang K et al. (2015). The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature 525(7567), 56-61.