Murine No Compensation Panels
Murine peripheral blood panels
The no compensation panels below have been optimized for murine peripheral blood and secondary lymphoid tissue such as spleen, bone marrow and lymph nodes, to help identify common cell subsets. Useful to create a simple four color panel or a great starting point when designing larger and more complex panels.These panels have been optimized by titrating the antibody to give the best signal resolution.
-
General identification panels
To identify T, B, NK and myeloid cells. -
Lymphocyte panels
To identify T and B cell populations such as naïve, memory and activated as well as helper and cytotoxic T cells. -
NK cell and myeloid panels
To identify NK and NKT cells and various myeloid populations.
General Identification Panels

T, B and myeloid cells
Markers |
Formats |
Catalog # |
Dilution |
|---|---|---|---|
|
CD3 |
Pacific Blue |
Neat |
|
|
CD19 |
Alexa Fluor 647 |
1:10 |
|
|
CD11b |
RPE-Alexa Fluor 750 |
1:10 |
|
|
Ly6G |
FITC |
1:10 |
Cells |
Surface Markers |
|---|---|
|
T cells |
CD3+ CD11b- CD19- Ly6G- |
|
B cells |
CD3- CD11b- CD19+ Ly6G- |
|
Monocytes/Macrophages |
CD3- CD11b+ CD19- Ly6G- |
|
Granulocytes |
CD3- CD11b+ CD19- Ly6G+ |
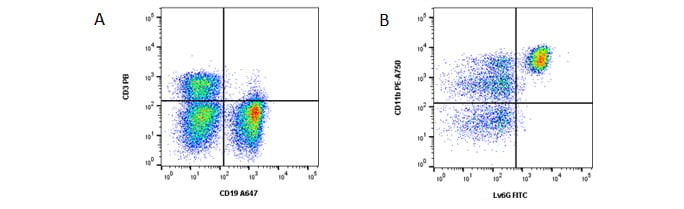
Fig. 1. General panel. A, identification of T and B cells. B, identification of myeloid cells from the CD3- CD19- population. Data acquired on the ZE5TM Cell Analyzer.

T, B, NK and myeloid cells
Markers |
Formats |
Catalog # |
Dilution |
|---|---|---|---|
|
CD3 |
Pacific Blue |
Neat |
|
|
B220 |
RPE-Alexa Fluor 750 |
1:10 |
|
|
CD49b |
FITC |
1:5 |
|
|
CD11b |
Alexa Fluor 647 |
1:10 |
Cells |
Surface Markers |
|---|---|
|
T cells |
CD3+ CD19- CD49- CD11b- |
|
B cells |
CD3- CD19+ CD49- CD11b- |
|
NK cells |
CD3- CD19- CD49b+ CD11b-/+ |
|
Myeloid |
CD3- CD19- CD49b- CD11b+ |
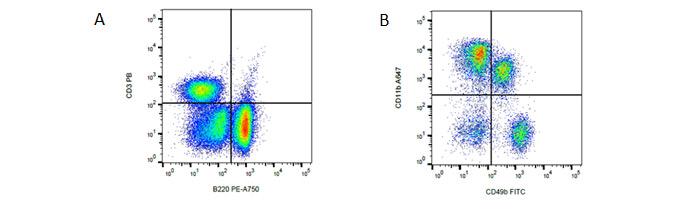
Fig. 2. General panel. A, identification of T and B cells. B, identification of NK cells and myeloid cells using CD3, B220, CD49b and CD11b. Data acquired on the ZE5 Cell Analyzer.
Lymphocyte Panels

T cells
Markers |
Formats |
Catalog # |
Dilution |
|---|---|---|---|
|
CD3 |
Pacific Blue |
Neat |
|
|
CD8 |
RPE-Alexa Fluor 750 |
1:5 |
|
|
CD4 |
Alexa Fluor 647 |
1:2 |
|
|
CD45 |
FITC |
1:5 |
Cells |
Surface Markers |
|---|---|
|
T helper |
CD45+ CD3+ CD4+ CD8- |
|
T cytotoxic |
CD45+ CD3+ CD8+ CD4- |
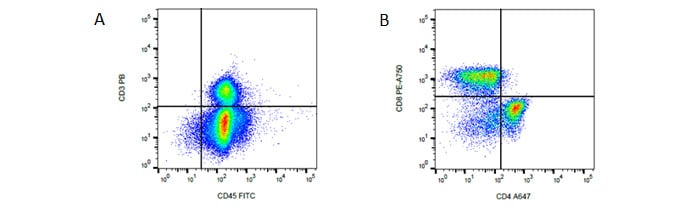
Fig. 3. T cell panel. A, identification of T cells. B, identification of T helper and T cytotoxic cells using CD45, CD3, CD4 and CD8. Data acquired on the ZE5 Cell Analyzer.
Incorporating CD44 and CD62L into this panel will allow identification of naïve, memory and effector T cells for both the helper and cytotoxic populations.
Cells |
Surface Markers |
|---|---|
|
Naive T helper cells |
CD3+ CD4+ CD62L+ CD44- |
|
Memory T helper cells |
CD3+ CD4+ CD62L-CD44+ |
|
Effector T helper cells |
CD3+ CD4+ CD62L- CD44mid |
|
Naïve T cytotoxic cells |
CD3+ CD8+ CD62L+ CD44- |
|
Memory T cytotoxic cells |
D3+ CD8+ CD62L- CD44+ |
|
Effector T cytotoxic cells |
CD3+ CD8+ CD62L- CD44mid |

Activated T cells
Markers |
Formats |
Catalog # |
Dilution |
|---|---|---|---|
|
CD3 |
Pacific Blue |
Neat |
|
|
CD4 or CD8 |
Alexa Fluor 647 |
1:2 or 1:5 |
|
|
B220 |
RPE-Alexa Fluor 750 |
1:10 |
|
|
CD69 |
FITC |
1:2 |
Cells |
Surface Markers |
|---|---|
|
Resting T helper cells |
CD3+ B220- CD4+ CD69- |
|
Activated T helper cells |
CD3+ B220- CD4+ CD69+ |
|
Resting T cytotoxic cells |
CD3+ B220- CD8+ CD69- |
|
Activated T cytotoxic cells |
CD3+ B220- CD8+ CD69+ |

B cells
Markers |
Formats |
Catalog # |
Dilution |
|---|---|---|---|
|
CD19 |
Pacific Blue |
1:10 |
|
|
B220 |
RPE-Alexa Fluor 750 |
1:10 |
|
|
IgM |
FITC |
|
|
|
CD23 |
Alexa Fluor 647 |
|
Cells |
Surface Markers |
|---|---|
|
Immature B cells |
CD19+ B220+ IgM+ CD23- |
|
Mature B cells |
CD19+ B220+ IgM- CD23+ |
B cell maturation in the blood and bone marrow can also be defined using IgM and IgD with immature B cells IgM + IgD- and mature B cells IgM-IgD + . Other markers can be used such as CD93 and CD21.

Naïve and memory B cells
Markers |
Formats |
Catalog # |
Dilution |
|---|---|---|---|
|
CD19 |
PB |
1:10 |
|
|
B220 |
RPE-Alexa Fluor 750 |
1:10 |
|
|
CD23 |
Alexa Fluor 647 |
Neat |
|
|
CD80 |
Alexa Fluor 647 |
1:5 |
Cells |
Surface Markers |
|---|---|
|
Naïve B cells |
CD19+ B220+ CD23+ CD80- |
|
Memory B cells |
CD19+ B220+ CD23- CD80+ |
The panel above is a simplification of the staining used to identify naïve and memory B cells, as the expression patterns of these surface markers can be variable compared to CD27 + and IgD expression in humans, which are more defined markers for naïve and memory B cells. Whilst this panel gives an indication of the amount of naïve and memory B cells, other markers could be included for more conclusive identification of these cells and other populations such as marginal zone B cells, depending upon the tissue being stained. These markers include CD38, CD62L, CD73, CD21 and PD-L2.
NK cell and Myeloid Panels

NK and NKT cells
Markers |
Formats |
Catalog # |
Dilution |
|---|---|---|---|
|
CD3 |
Pacific Blue |
Neat |
|
|
CD335 |
RPE-Cy7 |
Neat |
|
|
CD45 |
Alexa Fluor 647 |
1:10 |
|
|
CD122 |
FITC |
|
|
Cells |
Surface Markers |
|---|---|
|
NK precursors |
CD45+ CD3- CD122+ CD335- |
|
NK cells |
CD45+ CD3- CD122+ CD335+ |
|
NKT cells |
CD45+ CD3+ CD122+ CD335+ |
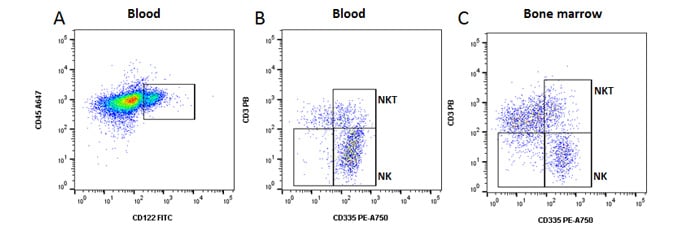
Fig 4. NK, NKT and NK precursor cells. A, identification of NK lineage cells using CD122. B, identification of CD335+ NK and CD335 + CD3 + NKT cells in blood. C, CD122 + CD335- NK cell precursors can also be seen in bone marrow. Data acquired on the ZE5 Cell Analyzer.

Myeloid Cell Panel
Markers |
Formats |
Catalog # |
Dilution |
|---|---|---|---|
|
CD11b |
Pacific Blue |
1:10 |
|
|
CD11c |
RPE-Alexa Fluor 750 |
1:2 |
|
|
Ly6C |
Alexa Fluor 647 |
1:10 |
|
|
Ly6G |
FITC |
1:10 |
Cells |
Surface Markers |
|---|---|
|
Granulocytes |
CD3- CD11b+ CD11c- Ly6G+ |
|
Classical Monocytes |
Ly6Chi CD11b+ CD11c- Ly6G- |
|
Non-Classical monocytes |
Ly6Clo CD11b+ CD11c- Ly6G- |
|
Dendritic cells |
CD11c+ CD11b+ Ly6C+/- Ly6G+/- |
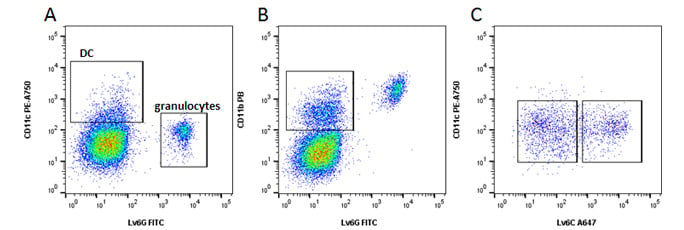
Fig 5. Myeloid cells. A, identification of dendritic cells (DC) and granulocytes. B, monocytes. C, Classical Ly6Chi and non-classical Ly6Clo monocytes. Data acquired on the ZE5 Cell Analyzer.
Although these no compensation panels are not available as pre-mixed four color panels cocktails, they have been optimized on the ZE5 Cell Analyzer and titrated for optimal resolution. To purchase pre-mixed panels for human, mouse, rat or dog cell surface markers, visit our antibody panels and cocktails webpage. If you can’t find the antibody you are interested in, you can search our database of over 3000 flow cytometry tested antibodies. To help you analyze your samples, information on the proportion of these populations in murine blood and tissues can be found on our cell frequency page.
When performing multicolor flow cytometry, you should consider careful sample preparation, Fc blocking, inclusion of a live/dead dye, doublet exclusion and using the appropriate controls. For more information, watch our flow cytometry webinar Optimize your Flow Cytometry or visit our application resources page.









