Autophagy Overview

- On This Page
- What is it
- Poster
- Detection
- Related genes
- Forms
- Stages
- Induction
- Formations
- Fusion
- Summary
What is Autophagy
Autophagy is an evolutionarily conserved physiological process by which cellular components (cargo) are recycled or degraded in lysosomes. It plays a critical role in maintaining cellular homeostasis by protein degradation and turnover of damaged cell organelles. Under conditions of minimal cell stress, autophagy rates are low, however autophagy is induced under stressful conditions such as in a tumor microenvironment or during nutrient starvation. Autophagy has a complex dual role in cell survival and cell death and has been rigorously studied in relation to the immune system and cancer. Although crucial to the maintenance of a healthy cell, under certain circumstances autophagy-dependent cell death (ADCD) can occur, which is a form of regulated cell death (Denton and Kumar, 2018), (Bialik et al. 2018).
View All Autophagy Detection Kits and AntibodiesAutophagy Detection
Autophagy plays a major role in the modulation of immune responses but also in malignancy protection and progression. Unlike some regulated cell death pathways, like apoptosis, autophagy can be difficult to detect and measure, as suitable reagents are limited. It is important to choose the most appropriate marker and application. For more information on detecting autophagy and best practices on autophagy detection by flow cytometry, select the appropriate link below:
Autophagy Related Genes
Yeast studies during the 1990s uncovered several important genes, which in 2003 were named autophagy-related genes (ATGs) (Klionsky 2003). Over 40 ATGs have since been discovered in yeast, many of which are conserved in animals and plants.
Research into autophagy has accelerated in recent years. A discovery made in 1999 connected autophagy to cancer, identifying downregulation of an autophagy gene, BECN1 (which encodes the protein Beclin 1), in breast tumor cells (Liang 1999). The link between autophagy and cancer continues to be the main focus of autophagy research today but the role of autophagy in cancer is not straightforward. Autophagy induction may slow the development of tumors, and in some circumstances inhibition of autophagy limits the growth of established tumors and improves the response to cancer therapeutics (Amaravati et al. 2019). In 2016, the Nobel Prize in Physiology or Medicine was awarded to Yoshonori Ohsumi for his discoveries of the mechanisms for autophagy, bringing autophagy and its role in cellular processes to the scientific forefront.
Forms of Autophagy
Autophagy can occur in several forms depending on the induction signal, type of cellular components that have been destroyed (cargo), and how the cargo is delivered into the lysosome for degradation, these include:
-
Macroautophagy – bulk degradation of nonselective cytoplasmic cargo; this is the most studied and best known autophagy pathway. It is important for recycling of damaged cell organelles and unused proteins and is upregulated under stress conditions
-
Microautophagy – direct lysosomal engulfment of cytoplasmic cargo. This is the second type of nonselective autophagy and it provides the cell with essential amino acids and nutrients in stress conditions. In addition, it also delivers glycogen into lysosomes
-
Chaperone-mediated autophagy – selective autophagy of target proteins. Proteins are trafficked across the lysosome membrane in complex with chaperone proteins and degraded upon recognition by a lysosomal membrane receptor, lysosomal-associated membrane protein 2A (LAMP-2A)
-
Mitophagy – selective autophagy of the mitochondria. This occurs to remove excessive or defective mitochondria which serves to prevent an accumulation of dysfunctional mitochondria in a cell
Other types of selective autophagy include golgiphagy, ribophagy, aggrephagy, ER-phagy, and lipophagy (Gatica 2018).
This mini-review covers the mechanisms of macroautophagy (hereafter referred to as autophagy) and the key markers involved in this cellular process.
Stages of Autophagy
Autophagy can be divided into five main stages: (Figure 1).
- Stress signals initiate formation of a double membrane structure called a phagophore.
- Phagophore elongation and engulfment of cytosolic cargo.
- The phagophore seals to form a spherical autophagosome.
- Autophagosome fusion with a lysosome.
-
Degradation of the contents of the autophagosome by proteolytic enzymes contained within the lysosome.
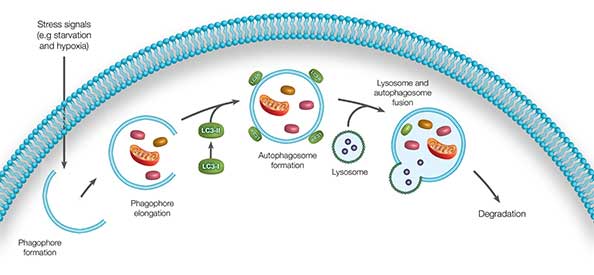
Fig. 1. Autophagy overview. The five main stages in the autophagy process.
Autophagy Induction
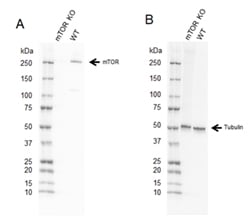
Fig. 2. Western blot analysis of mTOR CRISPR knockout HEK293 (mTOR KO) and wild type HEK293 (WT) whole cell lysates.
A, Rabbit Anti-mTOR Antibody (VPA00174) followed by detection with HRP conjugated Goat Anti-Rabbit IgG (H/L) (1/10,000 STAR208P) and B, hFAB Rhodamine Anti-Tubulin Primary Antibody (12004166) and visualized on the ChemiDoc MP Imaging System with a A, 37 s and B, 30 s exposure.
Autophagy normally occurs at low basal levels to maintain homeostasis, but is rapidly upregulated when cells require energy and nutrients, during high bioenergetic demand, structural remodeling, and to remove damaged cytoplasmic components. Autophagy needs to be carefully controlled as too much or too little can be detrimental to the cell. Autophagy is controlled by complex signaling pathways, key mediators for these are the Unc-51, like autophagy activating kinase (ULK1/2) complex and phosphoinositide 3-kinase class III (PI3K).
ULK1, a homolog of yeast Atg1, is a master nutrient sensor regulated by mammalian target of rapamycin complex (mTORC1) and 5’-AMP-activated protein kinase (AMPK). The PI3K and AKT (protein kinase B) signaling pathway regulates mammalian target of rapamycin (mTOR), a key component of mTORC1.
The PI3K pathway is activated under nutrient-rich conditions, resulting in mTORC1 activation and inhibition of autophagy by phosphorylating ULK1/2 at inhibiting sites. During starvation, AMPK levels increase. This results in inhibition of mTORC1 and phosphorylation of ULK1/2 at activating sites (Kim and Guan 2015). Downstream activation of Beclin 1 leads to phagophore formation and ultimately autophagy induction. This pathway is summarized in Figure 3.
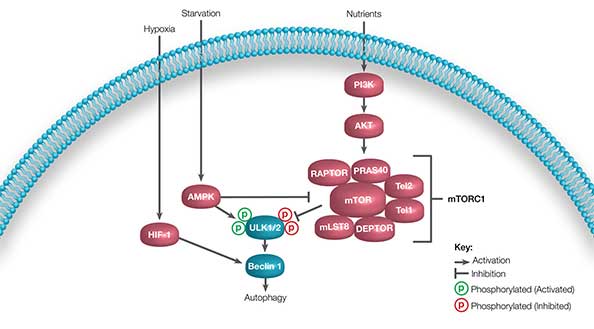
Fig. 3. Simplified major pathways of autophagy induction. In nutrient rich conditions, signaling occurs via PI3K and mTORC1 phosphorylates ULK1/2 at inhibiting sites (red P). During starvation, signaling via the AMPK pathway phosphorylates ULK1/2 at activating sites (green P) leading to Beclin 1 activation and autophagy activation. HIF-1 signaling induced by hypoxia can active Beclin 1 directly.
Autophagy can also be induced under cellular stress independent of mTORC1 (Figure 2). One example is HIF-1 activation of Beclin 1 under hypoxic conditions. In addition, eukaryotic initiation factor 2α (eIF2α) and mitogen-activated protein kinase 14 (MAPK14) are involved in the induction of autophagy under starvation conditions, DNA double-strand breaks and endoplasmic reticulum (ER) stress.
For more information on the role of mTOR in mTORC1 and mTORC2 signaling visit our mTOR Signaling Pathway page.
Phagophore and Autophagsome Formation
Autophagy proteins, including phosphorylated phosphatidylinositol (PI), assemble at membrane sites. These areas of membrane form a double membrane phagophore. The localized synthesis of phosphatidylinositol-3,4,5-triphosphate (PI3P) plays a major role in autophagosome assembly, by recruiting a number of effector proteins that lead to membrane remodeling. One member of this family of effector proteins are the homologs of yeast ATG-18 called WD-repeat protein interacting with phosphoinositides (WIPIs). There are four mammalian proteins -WIPI 1-4. WIPI2b recruits ATG16L1, which in turn recruits the ATG5-12 complex to form the larger ATG5/12/16L1 complex. This complex is required for ATG8 lipidation and has a role in elongation of the autophagsome. Other ATG proteins are also important for autophagosome formation including ATG3, ATG7, ATG8, ATG9,ATG13, ATG16, and ATG18.
Yeast only have one ATG8 gene, but in mammalian cells there are at least seven ATG8 paralogs which can be grouped into two categories:
- Gamma aminobutyric acid receptor associated proteins (GABARAP’s), GABARAP-L1/GEC1/ATG8L, GABARAP-L2/GATE-16, and GABARA-L3
-
MAP1LC3 (LC3) family: LC3A, LC3B, LC3B2, and LC3C
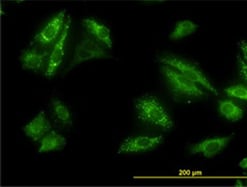
Fig. 4. Detection of p62 by immunofluorescence. Staining of HeLa cells with Mouse Anti-Human p62 (SQSTM1) Antibody (MCA4265Z).
GABARAP/GATE16 is involved in autophagosome sealing whereas LC3 is required for autophagosome expansion. Cytosolic forms of LC3 (LC3-I) are lipidated by conjugation to phosphatidylethanolamine (PE) to form a LC3-phosphatidylethanolamine conjugate (LC3-II), which is recruited to autophagosomal membranes. LC3-II is widely used as a marker of autophagosomes as it is the only protein that remains attached to the autophagosome following its completion, and is therefore a crucial marker when detecting autophagy (Mannack 2015).
p62 is a protein involved in selective autophagy and is also used as a marker of autophagy (Figure 4). It interacts via its C-terminal to ubiquitin or polyubiquitin chains on cargo to LC3 via a specific sequence motif resulting in the delivery of cargo to be degraded to the autophagosome. p62 accumulates when autophagy is inhibited and decreases under conditions of autophagy induction, as p62 is degraded along with the cargo in autolysosomes.
Fusion with Lysosomes and Degradation
Autophagosomes fuse with early and late endosomes as they travel along the microtubule network towards the lysosome, to form an amphisome. The structure acidifies as it matures due to the fusion and the actions of proton pumps on its outer membrane. It is thought that this acidification allows more efficient degradation of its engulfed contents. It then fuses with a lysosome to form an autolysosome, this fusion is dependent on recognition of phosphoinostide PI(3,5)P2 on the autophagosome membrane. Lysosomal membrane proteins, key to the fusion process, include LAMP1 and LAMP2, and the fusion event is controlled by soluble N-ethylmaleimide-sensitive factor attachment protein receptors (SNARE). An important SNARE protein on the autophagosome is syntaxin17 which forms a complex with two additional SNARE proteins - vesicle associated membrane protein 8 (VAMP8) on the lysosome and synaptosomal-associated protein 29 (SNAP29) in the cytoplasm. Ras–associated binding (Rab) proteins on membranes are also critical for fusion as they act as bridges to bring the compartments together (Nakamura and Yoshimori 2017). This final step in the autophagy process occurs when the contents of the autolysosome are degraded by lysosomal proteases such as cathepsins.
Summary
Autophagy is a cellular process that removes and recycles contents of the cell. This process occurs at a basal level, but is increased during cellular stress. There are several forms of autophagy, here we have described macroautophagy. It is a multistep process leading to the formation of autophagosomes which fuse with lysosomes to degrade the cellular content for recycling. To find out more about effective detection of autophagy using WB, IF, and IHC, best practice for autophagy detection by flow cytometry, and the role of autophagy in the immune system and cancer, select the appropriate link below:
For more information on all 13 of the Regulated Cell Death pathways, including available reagents, best practice for detection posters, and mini-reviews, visit our Regulated Cell Death webpage.
References
- Amaravadi R et al. (2019). Targeting autophagy in cancer: recent advances and future directions. Cancer Discovery 9, 1169–1181.
- Bialik S et al. (2018). Autophagy-dependent cell death – where, how and why a cell eats itself to death. J Cell Science. 131.
- Denton D and Kumar S (2019). Autophagy-dependent cell death. Cell Death Differ 26, 605–616.
- Gatica D et al. (2018). Cargo recognition and degradation by selective autophagy. Nat Cell Bio. 20 (3), 233-242.
- Kim TC and Guan KL (2015). mTOR: a pharmacologic target for autophagy regulation. J Clin Invest 125, 25–32.
- Klionsky DJ et al. (2003). A unified nomenclature for yeast autophagy-related genes. Dev Cell 5; 539–545.
- Liang XH et al. (1999). Induction of autophagy and inhibition of tumorigenesis by beclin 2. Nature 402 (6762), 672–6.
- Mannack LVJ and Lane JD (2015). The autophagosome: current understating of formation and maturation. Research and Reports in Biochemistry 5, 39–58.
- Nakamura S and Yoshimori T (2017). New insights into autophagosome-lysosome fusion. Journal of Cell Science 130 (7), 1209–1216.