Antibodies for Epigenetic Research

- On This Page
- DNA Methylation
- Histone Modifications
- RNA-Associated Silencing
- Epigenetics and Disease
- Epigenetics Range
- References
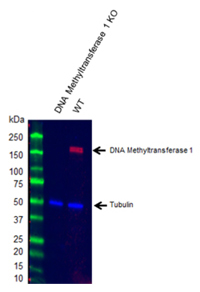
Fig.1. Western blot analysis of DNA methyltransferase 1 knockout 293T (KO) and wild type (WT) whole cell lysates probed with Rabbit Anti-DNA Methyltransferase 1 Antibody (VPA00026).
Epigenetics investigates heritable changes in gene expression without changes to the DNA sequence. Epigenetic mechanisms, such as DNA methylation, chromatin remodeling, and noncoding RNAs, regulate cell growth, development, and differentiation (Bird 2007).
DNA Methylation
DNA methylation is the addition of a methyl group to a cytosine nucleotide particularly within the CpG dinucleotide. The methylation of CpG sites is performed by DNA methyltransferases (DNMTs) (Egger et al. 2004). The canonical DNMT enzymes are DNMT1 (Figure 1), DNMT3A, and DNMT3B (Bestor 2000). DNA methylation regulates transcription, chromatin structure, DNA repair, and X chromosome inactivation. It is also important for the developmental control of gene expression (Robertson 2001 and 2002).
Histone Modifications
Histones are a family of proteins that are found in the nucleus and function to package DNA into nucleosomes. The amino-terminal tail domains of histones can be modified through acetylation and methylation. Histone acetylation usually denotes active chromatin, whereas deacetylation is found in transcriptionally inactive regions. Histone methylation marks both active and inactive regions of chromatin (Egger et al. 2004). Histone modifications also include arginine citrullination and Ser/Thr/Tyr phosphorylation. The majority of histone alterations control DNA transcription (Fardi et al. 2018).
RNA-Associated Silencing
MicroRNAs (miRNAs), small interfering RNAs (siRNAs), Piwi-interacting RNAs (piRNAs), and long noncoding RNAs (lncRNAs) are all epigenetic related noncoding RNAs (Cheng et al. 2019). Noncoding RNAs regulate gene expression and genome stability (Holoch and Moazed 2015).
Epigenetics and Disease
Tumor cells have different DNA methylation patterns compared to normal cells (Goelz et al. 1985). In cancer cells, hypomethylation in the DNA and hypermethylation at other sites are observed. Hypomethylation can cause expression of oncogenes whereas hypermethylation results in inhibition of tumor suppressor genes (Fardi et al. 2018). CpG islands, regions of the genome with a high number of CpG dinucleotide repeats, are extensively methylated in cancer cells (Robertson 2001 and 2002). Aberrant promoter methylation is linked to a loss of gene function. For example, silencing of BRCA1 by promoter hypermethylation occurs in sporadic variants of breast and ovarian cancer (Esteller et al. 2000). The methylation of EN1 has been found to be elevated in human salivary gland cystic carcinoma (Achim et al. 2011). Methylation levels have been demonstrated to be positively related to tumor size (Christensen et al. 2010).
Inhibition of histone/protein deacetylases (HDAC) has been shown in clinical and experimental investigations to have major anti-neoplastic effects via cytotoxic and proapoptotic pathways (Akimova T et al. 2012). Rett, ATRX, and fragile X syndromes are caused by naturally occurring mutations in genes that affect DNA methylation patterns (Robertson and Wolffe 2000). Epigenetic dysregulation also has a direct impact on the development of autoimmunity (Moosavi et al. 2016). Particularly, DNA methylation plays a significant role in autoimmune disorders by modifying gene expression profiles (Chen et al. 2016; Paul et al. 2016).
Epigenetic changes may potentially predict clinical outcomes. Large-scale genome-wide DNA methylation profiling revealed that specific DNA methylation patterns are characteristic of different forms of acute myeloid leukemia (Figueroa et al. 2010). Histone lysine methylation can be used as a key indicator of prostate cancer recurrence (Ellinger et al. 2010).
Due to the reversible nature of the epigenetic modifications, various epigenetic therapeutic approaches have emerged. The FDA approved the inhibition of DNMTs and HDACs in cancer treatment (Fardi et al. 2018).
Bio-Rad’s Antibody Range to Study Epigenetics
Browse Bio-Rad’s range of antibodies against epigenetics factors. Use the filters in the table below to sort the attributes and find the antibody that meets your needs. If you require any further assistance, please contact us.
Our Epigenetic Research Antibody Range
| Description | Target | Format | Clone | Applications | Citations | Code |
|---|
References:
- Achim B et al. (2011). CpG island methylation profiling in human salivary gland adenoid cystic carcinoma. Cancer 117, 2,898–2,909.
- Akimova T (2012). Histone/protein deacetylases and T-cell immune responses. Blood 119, 2,443–2,451.
- Bestor T (2000). The DNA methyltransferases of mammals. Hum Mol Genet. 9, 2,395–2,402.
- Bird A (2007). Perceptions of epigenetics. Nature 447, 396–398.
- Chen et al. (2016). Epigenomic profiling reveals an association between persistence of DNA methylation and metabolic memory in the DCCT/EDIC type 1 diabetes cohort. Proc Natl Acad Sci USA 113, E3002-11.
- Cheng Y et al. (2019). Targeting epigenetic regulators for cancer therapy: mechanisms and advances in clinical trials. Signal Transduct Target Ther 4, 62.
- Christensen B et al. (2010). Breast cancer DNA methylation profiles are associated with tumor size and alcohol and folate intake. PLOS Genet 6. Accessed December 14, 2021.
- Egger G et al. (2004). Epigenetics in human disease and prospects for epigenetic therapy. Nature 429. 457–463.
- Ellinger J et al. (2010). Global levels of histone modifications predict prostate cancer recurrence. Prostate 70, 61–69.
- Esteller M et al. (2000). Promoter hypermethylation and BRCA1 inactivation in sporadic breast and ovarian tumors. J Natl Cancer Inst. 92, 564-569.
- Fardi M et al. (2018). Epigenetic mechanisms as a new approach in cancer treatment: An updated review. Genes & Diseases 5, 304–311.
- Figueroa M et al. (2010). DNA Methylation signatures identify biologically distinct subtypes in acute myeloid leukemia. Cancer Cell 1, 13–27.
- Goelz S et al. (1985). Hypomethylation of DNA from benign and malignant human colon neoplasms. Science 228, 187–90.
- Holoch D and Moazed D (2015). RNA-mediated epigenetic regulation of gene expression. Nat Rev Genet 16, 71-84.
- Moosavi A et al. (2016). Role of epigenetics in biology and human diseases. Iran Biomed J. 20, 246–258.
- Paul D et al. (2016). Increased DNA methylation variability in type 1 diabetes across three immune effector cell types. Nat Commun 29. Accessed December 14, 2021.
- Robertson K. (2001). DNA methylation, methyltransferases, and cancer. Oncogene 20, 3,139–3,155.
- Robertson K (2002). DNA methylation and chromatin – unraveling the tangled web. Oncogene 21, 5,361–5,379.
- Robertson K and Wolffe A (2000). DNA methylation in health and disease. Nat Rev Genet 1, 11–19.




