Mini-review: Understanding the Eukaryotic Cell Cycle
Biological and Experimental Overview
The eukaryotic cell cycle is an evolutionarily conserved process that results in the replication of cells. It is tightly regulated, and includes three major checkpoints: G1, G2/M, and spindle (M). These checkpoints monitor the order, fidelity, and integrity of each phase of the cell cycle. For example, the G2/M checkpoint detects potential DNA damage, thus allowing repair before cell division. Defects in cell cycle progression often result in diseases such as cancer. Accordingly, this essential life cycle is routinely used to assess cell health, as well as for cancer prognosis and diagnosis. This mini-review provides an overview of the eukaryotic cell cycle, including common molecular techniques for evaluating proliferating cells.
Contents
1. Overview of the Cell Cycle in Eukaryotes
1.1. Introduction to the Eukaryotic Cell Cycle
1.2. Cell Cycle Control
1.3. DNA Replication in Eukaryotes
1.4. Key Cell Proliferation Biomarkers
2. Molecular Techniques for Assessing DNA Replication and Cell Cycle Kinetics
2.1. Assessing DNA Replication
2.2. Quantifying DNA Content
2.3. Measuring the Expression of Cyclins and Cdks
2.4. Detecting Cell Proliferation Markers
2.5. Summary and Future Outlook
1. Overview of the Cell Cycle in Eukaryotes
From as early as the 19th century, scientists have been intensely investigating the cell cycle. During this period, they discovered that new cells are derived from pre-existing cells (Nurse et al. 1998). However, the exact processes involved in cell division were largely unknown. Of particular scientific interest was to determine how the process differed among species.
In the last 60 years, there have been significant discoveries and insights into the molecular mechanisms involved in cell division in prokaryotes and eukaryotes. These studies demonstrated that the cell cycle in eukaryotes is much more complex than in prokaryotic organisms. Although, DNA replication and cell division occur in both cases, the processes vary significantly. A main difference lies in how these organisms replicate their DNA. Since the average eukaryotic cell has 25 times more DNA than prokaryotic cells, prokaryotic cell division does not include DNA condensation into chromosomes as observed in the eukaryotic cell cycle. Another major difference lies in the stages at which DNA replication occurs. Prokaryotes replicate their DNA continuously throughout their relatively short cell cycle whereas eukaryotic cells replicate DNA only in the S-phase of the cell cycle (discussed in detail in Section 1.2).
This mini-review will specifically discuss the intricacies of the eukaryotic cell cycle with special focus on DNA replication, cell cycle control, and key biomarkers of cell division.
1.1. Introduction to the Eukaryotic Cell Cycle
The highly regulated cell cycle is divided into phases, referred to as interphase (G1, S, and G2) and the mitotic (M) phase (Figure 1). In the gap 1 (G1) phase, the cell grows and acquires the energy needed for division. Cellular components, except for chromosomes, are duplicated at this stage. In the synthesis (S) phase, DNA replication occurs to duplicate the genetic material, with each chromosome now consisting of two sister chromatids. In the gap 2 (G2) phase, the cell prepares to divide by inducing metabolic changes that assemble the cytoplasmic components necessary for mitosis. During the M phase, nuclear division occurs, and the cell finally divides to create two identical daughter cells. The physical process of creating two daughter cells through division of the nucleus, cytoplasm, and plasma membrane is referred to as cytokinesis (derived from the Greek kyto or kýtos meaning container, body, or receptacle and kīnēsis meaning movement). At the end of cytokinesis each new cell consists of a full complement of DNA from the parent cell (Kapinas et al. 2013).
Another specialized cell division process known as meiosis is required to produce egg and sperm cells for reproduction. This process is split into meiosis I and meiosis II, in which meiosis I is unique to germ cells and meiosis II is similar to mitosis. However, in contrast to mitosis, the molecular and regulatory mechanisms involved in meiosis are less understood (Ohkura 2015).
Under certain conditions, a cell can exit the cell cycle and enter a state of quiescence referred to as the gap 0 (G0) phase. This phase is however reversible and G0 cells can return to the G1 phase and resume growth and division if appropriately stimulated.
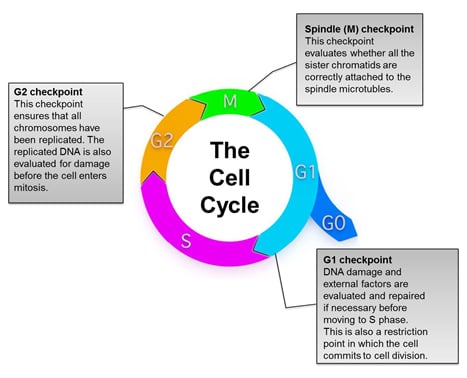
Fig. 1. Overview of the eukaryotic cell cycle. During cell division, cells pass through a series of stages collectively referred to as the cell cycle. To ensure that healthy cells are produced after each round of cell division, the cell cycle consists of three major checkpoints with distinct functions: G1, G2, and Spindle (M) checkpoints.
1.2. Cell Cycle Control
Each phase of the cell cycle is tightly regulated, with checkpoints in place near the end of G1, at the G2/M transition, and near the end of the metaphase stage of mitosis (spindle (M) checkpoint). These checkpoints are surveillance mechanisms whose function is to ensure that the generated daughter cells are duplicates of the parent cell complete with the accurate number of chromosomes and are mutation free (Figure 1). During the G1 checkpoint, cellular conditions necessary for progression through the cell cycle are evaluated. A cell generally passes the G1 checkpoint if it is an appropriate size, possesses adequate energy, and does not have damaged DNA. The main function of the G2 checkpoint is to ensure that replication of all chromosomes is complete and without introductions of mutations or unrepaired DNA damage. In addition, appropriate cell size and protein reserves are also assessed during this checkpoint. The spindle/M checkpoint ensures that all sister chromatids are correctly attached to the spindle microtubules and that each cell has the correct number of chromosomes.
These checkpoints halt cell cycle progression if the cell has not met each of the requirements being evaluated. This is necessary to allow the identified unfavorable conditions to be addressed. For example, detected DNA damage leads to the activation of the p53 transcription factor, which has been referred to as the ‘guardian of the genome’ due to its major role in maintaining genome stability (Lane 1992). The main function of p53 is to induce cell cycle arrest at the G1 or G2/M phases and initiate DNA repair. It activates gene expression of DNA repair genes such as P53R2 (Tanaka et al. 2000). p53 can also induce apoptosis as a last resort, if the damaged DNA cannot be repaired, by inducing expression of apoptotic genes such as BAX (Zilfou and Lowe 2009). Since it plays such an important role in preventing the continued cell cycle progression of cells with mutated DNA, p53 is considered a tumor suppressor (Zilfou and Lowe 2009). Consequently, it has been reported to be commonly mutated or absent in several types of cancer (Hussain and Harris 1998).
The master regulators of the cell cycle in eukaryotes are however heterodimeric enzyme complexes, which consist of cyclins and cyclin-dependent kinases (Cdks) (Murray 2004). The expression of cyclins increases or decreases in distinct phases of the cell cycle, and they are divided into groups based on the cell cycle phase that they regulate (Figure 2) (Murray 2004). However, in most cases, the concentration of Cdks remains relatively constant. Each Cdk subunit can associate with different cyclins, and the associated cyclin determines which protein substrates are phosphorylated by the Cdk-cyclin complex (Lodish et al. 2000). Moreover, Cdks have no kinase activity unless cyclin bound. In addition to the binding of cyclins, activation of the complex also requires phosphorylation of key residues in the activation loop of the Cdk subunit (Harper and Elledge 1998, Hochegger et al. 2008).
Several mechanisms have been identified for inhibiting activated cyclin-Cdk complexes. These include inhibitory phosphorylation of important residues such as tyrosine 15 and threonine 14 in Cdk1, degradation of the cyclin subunits by specific ubiquitin-mediated proteolysis, or association of the complex with a highly specific inhibitor protein such as p16 in the case of the cyclin D-Cdk4 complex (Hochegger et al. 2008, Kellogg 2003, Peters 2006, Serrano et al. 1993).
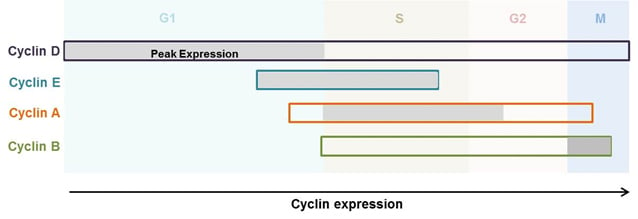
Fig. 2. Expression of cyclins throughout the cell cycle phases (Lodish et al. 2000). Cyclins are differentially expressed at various phases of the cell cycle and play distinct roles in cell cycle control. The figure demonstrates the stages in the cell cycle in which each cyclin is expressed. The grey shaded areas represent the peak expression of the respective cyclin.
The classical model of cell cycle control indicates that D-type cyclins and Cdk4 or Cdk6 regulate events in the early G1 phase (Nurse 2000). The cyclin E-Cdk2 complex then initiates the S phase and cyclin A-Cdk2 or cyclin A-Cdk1 complexes regulate the completion of the S phase. The cyclin B-Cdk1 complex is subsequently responsible for mitosis (Table 1). The transition between each phase of the cell cycle is mediated by protein phosphorylation, which is catalyzed by Cdks. In response, the removal of phosphate residues by phosphatases is also critical for cell cycle progression. For example, several phosphatases are involved in the control of mitosis (Chen et al. 2007). Protein phosphatase-2A1 (PP2A1) mediates the main phosphatase activity towards mitotic substrates (Sola et al. 1991). PP2A is deactivated when cells enter mitosis but is reactivated after the proteolysis of mitotic cyclins (Sola et al. 1991).
A decade ago, a revised model of eukaryotic cell cycle regulation, called the minimal threshold model, was proposed (Hochegger et al. 2008). This model stipulates that either Cdk1 or Cdk2 bound to cyclin A is sufficient to control all stages of interphase, whereas the cyclin B-Cdk-1 complex is necessary for the transition to mitosis (Hochegger et al. 2008). It also postulates that the differences between interphase and mitotic Cdks are not necessarily related to the specific cyclins they interact with. However, this is due to localization and a higher activity threshold for mitosis than interphase (Hochegger et al. 2008).
Knock out mice provide data in support of this model demonstrating that deletion of certain Cdks and cyclins does not lead to disruption of the cell cycle in somatic cells (Berthet and Kaldis 2007, Hochegger et al. 2008, Malumbres and Barbacid 2005). For example, the embryos of mice lacking Cdk2, Cdk4, and Cdk6 still carry out a functional cell cycle (Santamaria et al. 2007). However, according to the classical view, cells from these mice should not be able to progress beyond the G1 phase (Hochegger et al. 2008). This suggests that only a few complexes such as Cdk1-cyclin A are necessary for cell cycle progression. The minimal threshold model is also supported by other studies in yeast and mathematical modeling experiments (Coudreuse and Nurse 2010, Gérard et al. 2015). Further studies are however needed to confirm this proposed model, such as those that provide an explanation for the differential effects of Cdk deletions. According to Hochegger et al. (2008), this could be due to yet unknown kinase-independent functions of Cdk-cyclin complexes.
Table 1. Cyclins - key regulators of the cell cycle.
|
Cdk Binding Partner(s) of Cyclins |
Peak Expression in the Cell Cycle |
Role in the Cell Cycle |
|---|---|---|---|
|
Cyclin D |
Preferentially binds Cdk4 and Cdk6, can also bind Cdk1 and Cdk2 |
G1 |
Cyclin D1 is required for the transition from G0 to G1. Forms a complex with Cdk4 to activate the retinoblastoma protein, which subsequently upregulates cyclin E expression |
|
Cyclin E |
Preferentially binds Cdk2, can also bind Cdk1 |
G1/S |
The cyclin E-Cdk2 complex is required for S phase initiation |
|
Cyclin A |
Preferentially binds Cdk2, can also bind Cdk1 |
S/G2 |
Cyclin A together with Cdk1 or Cdk2 regulates the completion of the S phase and entry into mitosis |
|
Cyclin B |
Preferentially binds Cdk1, can also bind Cdk2 |
M |
Cyclins B1 and B2 interact with Cdk1 in the M phase to form the M phase/maturation-promoting factor (MPF), which regulates the processes that promote mitotic spindle assembly and finally, cell division |
Hochegger et al. (2008). Cdk, cyclin-dependent kinase.
1.3. DNA Replication in Eukaryotes
DNA replication is an integral part of the cell cycle that distinctly takes place in the S phase. This process must occur with exquisite accuracy prior to cell division (Alberts et al. 2002).
DNA replication begins with the unwinding of the DNA double helix by a member of the helicase enzyme family (Figure 3, step 1). Helicases unwind DNA by breaking the hydrogen bonds between nucleotide base pairs. Single stranded DNA binding proteins stabilize the unwound DNA and keep the strands from rejoining, creating a Y-shaped replication fork in which the synthesis of new DNA strands occurs (Figure 3, step 2).
At the replication fork, the separated DNA strands serve as a template that guides the insertion of complementary nucleotides to form new DNA. For example, a cytosine nucleotide on the template strand guides the insertion of a guanine on the new strand. Similarly, an adenine nucleotide guides the insertion of thymine.
Prior to the addition of the complementary nucleotides, primase adds RNA primers to the template strands, leaving a free 3′ OH group to initiate synthesis of the new strand. DNA polymerase ε then binds the template strand and elongates the RNA primer by adding nucleotides to the free 3′ end (Figure 3, step 3).
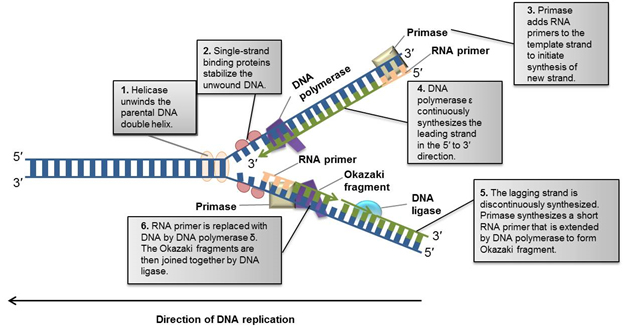
Fig. 3. Overview of DNA replication (Alberts et al. 2002). Before cells divide, they must replicate their DNA. This figure demonstrates the DNA replication fork in eukaryotes and the steps involved in generating a new complementary DNA strand.
DNA polymerase only functions in the 5′ to 3′ direction (see Figure 3 for DNA orientation). Consequently, only one new DNA strand (called the leading strand) can be continuously synthesized. Synthesis of the other strand (called the lagging strand) requires a more complex process (Figure 3, steps 5 and 6).
To create the lagging 3′ to 5′ strand, several RNA primers are discontinuously elongated by DNA polymerase in the 5′ to 3′ direction in segments called Okazaki fragments (Figure 3, step 5). Similar to synthesis of the leading strand, primase introduces RNA primers to initiate further elongation. RNase removes the RNA primers and another DNA polymerase (DNA polymerase δ) fills the gaps between the Okazaki fragments. DNA ligase I then fills nicks in both the leading and lagging strands to complete the newly synthesized DNA molecule (Figure 3, step 6).
At the completion of the DNA replication process, each daughter cell of an actively dividing parent cell inherits a new DNA double helix that contains one old and one new strand.
1.4. Key Cell Proliferation Biomarkers
In addition to cyclins and Cdks (described in Section 1.2), Ki-67, proliferation cell nuclear antigen (PCNA), and mini-chromosome maintenance protein 2 (MCM2) are also key proteins for regulating cellular growth and proliferation.
Ki-67 is a nuclear protein that is present during all phases of the cell cycle (late G1, S, G2, M) but is absent in resting cells (G0 phase) (Gerdes et al. 1984). Its protein expression is low during G1 and early S phase and gradually increases to reach a maximum during mitosis (Li et al. 2014). During mitosis, Ki-67 is specifically expressed on the surface of chromosomes, and is primarily involved in chromatin condensation (Cuylen et al. 2016, van Dierendonck et al. 1989). Ki-67 overexpression has been reported in many cancer types, including those of the breasts and lungs, and this is associated with reduced patient survival (Pollack et al. 2004, Shiba et al. 2000, Stuart-Harris et al. 2008). As a result, Ki-67 has been used in the clinic as a prognostic and predictive marker for certain cancer types such as breast cancer (Li et al. 2014). However as a clinical biomarker for cancer, its usefulness and clinical significance is challenged by conflicting studies demonstrating inter-laboratory and inter-observer variability (Albarracin and Dhamne 2014).
PCNA is an essential evolutionarily conserved protein; highlighted by the fact that knocking out PCNA in mice, results in embryonic lethality (Hu and Xiong 2006, Roa et al. 2008, Strzalka and Ziemienowicz 2011). The first role of the protein was the identification of PCNA’s involvement in DNA synthesis as an auxiliary protein for DNA polymerase δ (Bravo et al. 1987). Since then, our understanding of PCNA’s function has been expanded, and the protein has been reported to play a role in various essential cellular processes such as DNA repair, chromatin remodeling, chromosome segregation, and cell cycle progression (Strzalka and Ziemienowicz 2011). PCNA has been described as the ‘ringmaster of the genome’ due to these diverse roles (Paunesku et al. 2001). PCNA also impacts cell fate through its interaction with p53. The PCNA gene is induced by p53, which leads to DNA repair when PCNA is highly expressed. In the absence of the tumor suppressor, PCNA can induce DNA replication. Low or absent PCNA expression leads to apoptosis (Paunesku et al. 2001). Therefore, PCNA is a critical protein whose expression dictates cell fate (Paunesku et al. 2001). PCNA is primarily expressed in late G1 and S phases, decreases its expression in G2 and M phases, and is low or absent in G0 and early G1 phases (Kurki et al. 1986). Due to its role in replication and DNA repair, PCNA is considered a marker of cell proliferation in many cancers including cervical cancer and gliomas (Lv et al. 2016).
MCM2 is one of six members of the MCM protein family, which consists of MCM2-MCM7. These proteins play a key role in the initiation of DNA replication by forming the pre-replication complex (pre-RC) in the G1 phase (Maiorano et al. 2006). The complex is critical for DNA replication in the subsequent S phase as it also stimulates the unwinding of the parental DNA strands (Maiorano et al. 2006, Labib et al. 2000). Compared to the other MCM family members, MCM 2 is distinctly expressed throughout the cell cycle. MCM3-MCM7 proteins are not expressed in stoichiometric amounts and are regulated differently (Todorov et al. 1998). For example, MCM7 is more abundant in quiescent cells than in proliferating cells compared to MCM2 (Tsuruga et al. 1997). MCM2 is highly expressed in early G1, moderately expressed in S, G2, and M phases, and absent during G0 (Maiorano et al. 2006). In addition, MCM2 demonstrates distinct cellular localization patterns that can be measured in cycling cells. It is expressed in the nucleus throughout the cell cycle, but is tightly bound to chromatin during G1 (Todorov et al. 1995). It is then displaced in the S phase and remains unbound in the G2 and M phases (Todorov et al. 1995). The localization and expression pattern of MCM2 therefore makes it an ideal proliferation marker. It is also used as a diagnostic and prognostic marker in cancer types such as kidney cancer and gliomas (Giaginis et al. 2010, Todorov et al. 1998).
Table 2 summarizes the cell cycle expression, function, and diagnostic use of these key proliferation biomarkers in cancer.
Table 2. Key cell proliferation biomarker summary.
Proliferation Biomarker |
Expression during the Cell Cycle |
Function |
Application in Cancer Diagnostics |
|---|---|---|---|
|
G0: not expressed G1: low expression S: low expression G2: increased expression M: maximum expression |
Primarily involved in chromatin condensation during mitosis |
Used as a prognostic and predictive marker in cancer types such as breast and lung cancer
Inter-laboratory and inter-observer variations limit its clinical usefulness |
|
|
G0: not expressed G1: high expression in early G1 S: moderate expression G2: moderate expression M: moderate expression |
Forms part of pre-replicative complex to initiate DNA replication
Involved in unwinding parental DNA |
Used as a diagnostic and prognostic biomarker in several cancer types such as kidney cancer and gliomas |
|
|
G0: low expression or not expressed G1: high expression in late G1 S: high expression G2: reduced expression M: reduced expression |
Involved in DNA replication and repair, chromatin remodeling, chromosome segregation, and cell cycle progression |
Marker of cell proliferation in several cancer types including cervical cancer and gliomas |
MCM2, mini-chromosome maintenance protein 2; PCNA, proliferation cell nuclear antigen.
2. Molecular Techniques for Assessing DNA Replication and Cell Cycle Kinetics
Our current understanding of the cell cycle and the key proteins involved has led to the development of a number of methods for quantifying and evaluating cell cycle processes and cell growth. These techniques are routinely applied in cancer and cell biology research. The most commonly used methods to assess dividing cells are highlighted below.
2.1. Assessing DNA Replication
Measuring newly synthesized DNA is the most common method for detecting proliferating cells, as well as assessing individual cell cycle phases. Traditional methods utilize chemical compounds that are incorporated into DNA instead of certain nucleotides. Thymidine analogs, which are incorporated into newly synthesized DNA during DNA replication, are especially popular. Traditionally, 5′-bromo-2′-deoxyuridine (BrdU) has been used for measuring DNA synthesis. The incorporated BrdU is detected using specific antibodies that have minimal cross-reactivity to thymidine (Magaud et al. 1989). Although BrdU incorporation allows accurate detection of newly synthesized DNA, it has been reported to have negative genetic and molecular effects on the biological sample (Anda et al. 2014, Lehner et al. 2011). Therefore, experiments utilizing BrdU should include appropriate controls. Other thymidine analogs commonly utilized for measuring DNA replication include 5′-chloro-2′-deoxyuridine (CldU), 5′-iodo-2′-deoxyuridine (IdU), and 5′-ethynyl-2′-deoxyuridine (EdU) (Salic and Mitchison 2008, Tuttle et al. 2010).
2.2. Quantifying DNA Content
The amount of DNA in a cell can be quantified using fluorescent dyes that stoichiometrically bind DNA. Accordingly, the amount of dye detected is directly proportional to the amount of DNA present in the cell. These dyes are therefore suitable for discriminating between various phases of the cell cycle based on the alterations in DNA that occur. For instance, cells in the S phase of the cell cycle will have more DNA than cells in the G1 phase. Similarly, cells in the G2 phase will have twice as much DNA and therefore higher fluorescence readouts than G1 cells. Common total DNA binding dyes include propidium iodide (PI), 4′,6′-diamidino-2-phenylindole (DAPI), and 7-aminoactinomycin-D (7-AAD). The intensity of these dyes is generally measured by flow cytometry. To effectively bind DNA, the cell samples should be permeabilized, as these dyes cannot enter living cells with intact membranes. Furthermore, these dyes also bind RNA; therefore RNAse treatment is required prior to addition of the dye to ensure that only DNA content is measured.
2.3. Measuring the Expression of Cyclins and Cdks
The distinct peak expression of cyclins and Cdks at various phases of the cell cycle can be exploited for cell cycle analysis (Figure 2). The total levels of individual cyclins as well as their phosphorylated forms can be detected using immunodetection methods such as western blotting or enzyme-linked immunosorbent assay (ELISA). Combining these techniques with assessment of DNA synthesis provides a thorough analysis of the cell cycle.
2.4. Detecting Cell Proliferation Markers
Analysis of cell proliferation markers (Ki-67, PCNA, and MCM2) can be used to identify proliferating cells because of their distinct expression throughout the cell cycle (Table 2). Antibodies specific to these proteins allow the distinction between actively dividing cells (positive expression) and quiescent (G0) cells (low or negative expression). This method of identifying proliferating cells is most common in pathological analyses of tumor tissue using immunohistochemistry. However, it is important to highlight that the results obtained using this method are only indicative of the number of proliferating cells rather than a direct measurement of the proliferation rate.
2.5. Summary and Future Outlook
The cell cycle is an ordered process that results in the replication of cells. This process is evolutionarily conserved, and defects in cell cycle control often result in diseases such as cancer (Harashima et al. 2013). In contrast to the 1800s when cell division was first observed, there have been significant advances in our understanding of how cells divide, with the development of novel techniques to measure this process experimentally. The identification of the molecular markers implicated in cell growth and cancer (Table 2) have also led to their investigation as promising targets of cancer therapy. For instance, because Ki-67 is associated with aggressive tumor growth and poor prognosis, it is currently being evaluated as a therapeutic target for cancer therapy (Rahmanzadeh et al. 2010, Ricciardi et al. 2015a, Ricciardi et al. 2015b). Studies have shown that inhibition of Ki-67 either via antibody inhibitors or antisense oligonucleotides leads to decreased cell division, which further supports its utility as a target for effective cancer treatment (Kausch et al. 2003, Starborg et al. 1996, Zhang et al. 2007).
References
- Albarracin C and Dhamne S (2014). Ki67 as a biomarker of prognosis and prediction: Is it ready for use in routine pathology practice. Curr Breast Cancer Rep 6, 260-266.
- Alberts B et al. (2002). DNA replication mechanisms. In Molecular Biology of the Cell, 4th edition, (New York: Garland Science), ncbi.nlm.nih.gov/books/NBK26850/, accessed April 25, 2017.
- Anda S et al. (2014). Cell-cycle analyses using thymidine analogues in fission yeast. PloS One 9, e88629.
- Berthet C and Kaldis P (2007). Cell-specific responses to loss of cyclin-dependent kinases. Oncogene 26, 4469-4477.
- Bravo R et al. (1987). Cyclin/PCNA is the auxiliary protein of DNA polymerase-delta. Nature 326, 515-517.
- Chen F et al. (2007). Multiple protein phosphatases are required for mitosis in Drosophila. Curr Biol 17, 293-303.
- Coudreuse D and Nurse P (2010). Driving the cell cycle with a minimal CDK control network. Nature 468, 1074-1079.
- Cuylen S et al. (2016). Ki-67 acts as a biological surfactant to disperse mitotic chromosomes. Nature 535, 308-312.
- Gerdes J et al. (1984). Cell cycle analysis of a cell proliferation-associated nuclear antigen defined by the monoclonal antibody Ki-67. J Immunol 133, 1710-1715.
- Gérard C et al. (2015). Cell cycle control by a minimal Cdk Network. PLoS Comput Biol 11, e1004056.
- Giaginis C et al. (2010). MCM proteins as diagnostic and prognostic tumor markers in the clinical setting. Histol Histopathol 25, 351-370.
- Harashima H et al. (2013). Cell cycle control across the eukaryotic kingdom. Trends Cell Biol 23, 345-356.
- Harper JW and Elledge SJ (1998). The role of Cdk7 in CAK function, a retro-retrospective. Genes Dev 12, 285-289.
- Hochegger H et al. (2008). Cyclin-dependent kinases and cell-cycle transitions: does one fit all? Nat Rev Mol Cell Biol 9, 910-916.
- Hu J and Xiong Y (2006). An evolutionarily conserved function of proliferating cell nuclear antigen for Cdt1 degradation by the Cul4-Ddb1 ubiquitin ligase in response to DNA damage. J Biol Chem 281, 3753-3756.
- Hussain SP and Harris CC (1998). Molecular epidemiology of human cancer: Contribution of mutation spectra studies of tumor suppressor genes. Cancer Res 58, 4023-4037.
- Kapinas K et al. (2013). The abbreviated pluripotent cell cycle. J Cell Physiol 228, 9-20.
- Kausch I et al. (2003). Antisense treatment against Ki-67 mRNA inhibits proliferation and tumor growth in vitro and in vivo. Int J Cancer 105, 710-716.
- Kellogg DR (2003). Wee1-dependent mechanisms required for coordination of cell growth and cell division. J Cell Sci 116, 4883-4890.
- Kurki P et al. (1986). Expression of proliferating cell nuclear antigen (PCNA)/cyclin during the cell cycle. Exp Cell Res 166, 209-219.
- Labib K et al. (2000). Uninterrupted MCM2-7 function required for DNA replication fork progression. Science 288, 1643-1647.
- Lane DP (1992). Cancer. p53, guardian of the genome. Nature 358, 15-16.
- Lehner B et al. (2011). The dark side of BrdU in neural stem cell biology: detrimental effects on cell cycle, differentiation and survival. Cell Tissue Res 345, 313-328.
- Li TL et al. (2014). Ki-67 is a promising molecular target in the diagnosis of cancer (Review). Mol Med Rep 11, 1566-1572.
- Lodish H et al. (2000). Overview of the cell cycle and its control. In Molecular Cell Biology, 4th edition (New York: WH Freeman & Co.), ncbi.nlm.nih.gov/books/NBK21466/, accessed May 2, 2017.
- Lv Q et al. (2016). Proliferating cell nuclear antigen has an association with prognosis and risks factors of cancer patients: a systematic review. Mol Neurobiol 53, 6209-6217.
- Magaud JP et al. (1989). Double immunocytochemical labeling of cell and tissue samples with monoclonal anti-bromodeoxyuridine. J Histochem Cytochem 37, 1517-1527.
- Maiorano D et al. (2006). MCM proteins and DNA replication. Curr Opin Cell Biol 18, 130-136.
- Malumbres M and Barbacid M (2005). Mammalian cyclin-dependent kinases. Trends Biochem Sci 30, 630-641.
- Murray AW (2004). Recycling the cell cycle: cyclins revisited. Cell 116, 221-234.
- Nurse P et al. (1998). Understanding the cell cycle. Nat Med 4, 1103-1106.
- Nurse P (2000). A long twentieth century of the cell cycle and beyond. Cell 100, 71-78.
- Ohkura H (2015). Meiosis: an overview of key differences from mitosis. Cold Spring Harb Perspect Biol 7, pii: a015859.
- Paunesku T et al. (2001). Proliferating cell nuclear antigen (PCNA): ringmaster of the genome. Int J Radiat Biol 77, 1007-1021.
- Peters JM (2006). The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat Rev Mol Cell Biol 7, 644-656.
- Pollack A et al. (2004). Ki-67 staining is a strong predictor of distant metastasis and mortality for men with prostate cancer treated with radiotherapy plus androgen deprivation: Radiation therapy oncology group trial 92-02. J Clin Oncol 22, 2133-2140.
- Rahmanzadeh R et al. (2010). Ki-67 as a molecular target for therapy in an in vitro 3D model for ovarian cancer. Cancer Res 70, 9234-9242.
- Ricciardi GR et al. (2015a). Androgen receptor (AR), E-Cadherin, and Ki-67 as emerging targets and novel prognostic markers in triple negative breast cancer (TNBC) patients. PLoS One 10, e0132647.
- Ricciardi GR et al. (2015b). Correction: Androgen receptor (AR), E-Cadherin, and Ki-67 as emerging targets and novel prognostic markers in triple negative breast cancer (TNBC) patients. PLoS One 10, e0132647.
- Roa S et al. (2008). Ubiquitylated PCNA plays a role in somatic hypermutation and class switch recombination and is required for meiotic progression. Proc Natl Acad Sci USA 105, 16248-16253.
- Salic A and Mitchison TJ (2008). A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc Natl Acad Sci USA 105, 2415-2420.
- Santamaria D et al. (2007). Cdk1 is sufficient to drive the mammalian cell cycle. Nature 448, 811-815.
- Serrano M et al. (1993). A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature 366, 704-707.
- Shiba M et al. (2000). Ki-67 immunostaining and other prognostic factors including tobacco smoking in patients with resected nonsmall cell lung carcinoma. Cancer 89, 1457-1465.
- Sola MM et al. (1991). p34cdc2 phosphorylation sites in histone H1 are dephosphorylated by protein phosphatase 2A1. Biochim Biophys Acta 1094, 211-216.
- Starborg M et al. (1996). The murine Ki-67 cell proliferation antigen accumulates in the nucleolar and heterochromatic regions of interphase cells and at the periphery of the mitotic chromosomes in a process essential for cell cycle progression. J Cell Sci 109, 143-153.
- Strzalka W and Ziemienowicz A (2011). Proliferating cell nuclear antigen (PCNA): a key factor in DNA replication and cell cycle regulation. Ann Bot 107, 1127-1140.
- Stuart-Harris R et al. (2008). Proliferation markers and survival in early breast cancer: a systematic review and meta-analysis of 85 studies in 32,825 patients. Breast 17, 323-334.
- Tanaka H et al. (2000). A ribonucleotide reductase gene involved in a p53-dependent cell-cycle checkpoint for DNA damage. Nature 404, 42-49.
- Todorov IT et al. (1995). A human nuclear protein with sequence homology to a family of early S phase proteins is required for entry into S phase and for cell division. J Cell Sci 107, 253-265.
- Todorov IT et al. (1998). HsMCM2/BM28: a novel proliferation marker for human tumors and normal tissues. Lab Invest 78, 73-78.
- Tsuruga H et al. (1997). Expression, nuclear localization and interactions of human MCM/P1 proteins. Biochem Biophys Res Commun 236, 118-125.
- Tuttle AH et al. (2010). Immunofluorescent detection of two thymidine analogues (CIdU and IdU) in primary tissue. J Vis Exp 46, 2166.
- Van Dierendonck JH et al. (1989). Nuclear distribution of the Ki-67 antigen during the cell cycle: comparison with growth fraction in human breast cancer cells. Cancer Res 49, 2999-3006.
- Zhang P et al. (2007). Versatile photosensitizers for photodynamic therapy at infrared excitation. J Am Chem Soc 129, 4526-4527.
- Zilfou JT and Lowe SW (2009). Tumor suppressive functions of p53. Cold Spring Harb Perspect Biol 1, a001883.




