
Popular topics

-
References
Bernstock JD et al. (2017). Topotecan is a potent inhibitor of SUMOylation in glioblastoma multiforme and alters both cellular replication and metabolic programming. Sci Rep 7, 7425.
Bouchard C et al. (1998). Control of cell proliferation by Myc. Trends Cell Biol 8, 202-206.
Cremona CA et al. (2012). Sumoylation and the DNA Damage Response. Biomolecules 2, 376-388.
Eifler K and Vertegall ACO (2015). SUMOylation-mediated regulation of cell cycle progression and cancer. Trends Biochem Sci 40, 779-793.
González-Prieto R et al. (2015). c-Myc is targeted to the proteasome for degradation in a SUMOylation-dependent manner, regulated by PIAS1, SENP7 and RNF4. Cell Cycle 14, 1859-72.
Hendriks IA and Vertegaal ACO (2016). A comprehensive compilation of SUMO proteomics. Nat Rev Mol Cell Biol 17, 581-95.
Huang J et al. (2012). SUMO1 modification of PTEN regulates tumorigenesis by controlling its association with the plasma membrane. Nat Commun 3, 911.
Leslie NR and Longy M (2016). Inherited PTEN mutations and the prediction of phenotype. Semin Cell Dev Biol Vol 52, 30-38.
Li R et al. (2013). Akt SUMOylation regulates cell proliferation and tumorigenesis. Cancer Res 73, 5742-53.
Miller DM et al. (2012). c-Myc and Cancer Metabolism. Clin Cancer Res 18:5546-5553.
Myatt SS et al. (2014). SUMOylation inhibits FOXM1 activity and delays mitotic transition. Oncogene 33, 4316-4329.
Nandi D et al. (2017). FoxM1: Repurposing an oncogene as a biomarker. Semin Cancer Biol pii: S1044-579X(17)30061-5.
Schimmel J et al. (2014). Uncovering SUMOylation dynamics during cell-cycle progression reveals FoxM1 as a key mitotic SUMO target protein. Mol Cell 53, 1053-66.
Song MS et al. (2012). The functions and regulation of the PTEN tumour suppressor. Nature Rev Mol Cell Biol 13, 283-296.
Yang C et al. (2013). Inhibition of FOXM1 transcription factor suppresses cell proliferation and tumor growth of breast cancer. Cancer Gene Ther 20, 117-124.
Zona S et al. (2014). FOXM1: An emerging master regulator of DNA damage response and genotoxic agent resistance. Biochim Biophys Acta 1839:1316-1322.
SUMOylation: a Heavyweight in the Regulation of Cell Cycle Progression and Cell Proliferation – Part Two
In the second part of our series on SUMOylation, we examine how key cell signaling proteins are regulated by SUMOylation, the effect of their aberrant SUMOylation on signaling pathways and the potential of SUMO inhibitors in cancer therapy.
Regulation of Key Signaling Proteins by SUMOylation
To date, around 3,600 SUMOylated proteins have been identified, which include a large variety of tumor suppressors, (proto)-oncogenes, transcription factors and DNA damage and repair proteins (Hendriks and Vertegaal 2016, Cremona et al. 2012). Aberrant SUMOylation of these proteins can result in cell cycle and cell proliferation defects, and ultimately cancer development (Eifler and Vertegall 2015). The examples below of SUMOylated signaling proteins provide an overview of the impact of this modification and how misregulation can trigger cancer.
PTEN Phosphatase
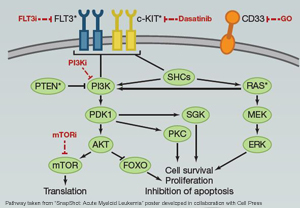 The phosphatase PTEN is a tumor suppressor, which blocks activation of the PI3K-AKT pathway by dephosphorylating PIP3 (Phosphatidylinositol (3,4,5)-trisphosphate) at the plasma membrane (Song at al. 2012). The gene is one of the most frequently mutated and deleted genes in human cancers (Leslie and Longy 2016) including melanoma, and Acute Myeloid Leukemia. Loss of PTEN function has been reported to promote both cell proliferation and survival (Leslie and Longy 2016). SUMOylation of PTEN is essential for its association with the plasma membrane. Aberrant SUMOylation results in increased PIP3 levels and therefore activation of the PI3K-AKT pathway, as well as increased cell proliferation (Huang et al. 2012).
The phosphatase PTEN is a tumor suppressor, which blocks activation of the PI3K-AKT pathway by dephosphorylating PIP3 (Phosphatidylinositol (3,4,5)-trisphosphate) at the plasma membrane (Song at al. 2012). The gene is one of the most frequently mutated and deleted genes in human cancers (Leslie and Longy 2016) including melanoma, and Acute Myeloid Leukemia. Loss of PTEN function has been reported to promote both cell proliferation and survival (Leslie and Longy 2016). SUMOylation of PTEN is essential for its association with the plasma membrane. Aberrant SUMOylation results in increased PIP3 levels and therefore activation of the PI3K-AKT pathway, as well as increased cell proliferation (Huang et al. 2012).
AKT1/PKB Kinase
The serine/threonine protein kinase AKT1 is a key regulator of cell proliferation and apoptosis. Li et al. (2013) showed that AKT SUMOylation results in activation of the kinase domain and promotes tumorigenesis. By systematically point-mutating lysine residues (K) the authors identified K267 as the major SUMOylation site. They showed that mutating K267 to arginine or alanine eliminated the AKT kinase activity, and reduced cell migration as well as cell proliferation (Li et al. 2013).
FOXM1 Transcription Factor
The forkhead box protein M1, FOXM1 transcription factor, is a “master regulator” of cell cycle gene expression and has a critical role in the G1 to S and G2 to M cell cycle phase transitions (Yang et al. 2013). In addition to this critical role, FOXM1 regulates expression of DNA damage and repair genes, which contribute to the maintenance of genome integrity (Zona et al. 2014). This feature has contributed to the oncogene classification of FOXM1 and has generated interest in the protein as a cancer biomarker (Nandi et al. 2017). Although there is consensus in the literature of FOXM1 SUMOylation , different SUMO isoform conjugations and opposing functional effects of these SUMOylation events have been reported.
Schimmel et al. (2014) showed evidence of FOXM1 SUMO-2 conjugation, which increased the activity of the transcription factor by blocking its dimerization and thereby lifting its autorepression. On the other hand, Myatt et al. (2014) found evidence of FOXM-1 SUMO-1 conjugation, which they reported to result in reduced transcription factor activity.
Myc Transcription Factor
The Myc/c-Myc transcription factor is a well characterized oncogene and a key regulator of cell proliferation (Bouchard et al. 1998). Myc gene mutations as well as aberrant gene expression induced by oncogenic signals frequently result in Myc overexpression, which has been reported in various cancer types (Miller et al. 2012). SUMOylation plays a critical role in the regulation of Myc protein levels. It also targets Myc proteins for ubiquitination and subsequent degradation by the 26S proteasome (González-Prieto et al. 2015).
SUMO Conjugation Components as Targets for Cancer Therapy
SUMOylation is important for transcriptional regulation of oncogene expression, cell cycle progression and cell proliferation. This makes the SUMO pathway, like the ubiquitin pathway, an attractive target for cancer therapy. SUMOylation has also been associated with maintaining genome integrity and key components of the SUMO conjugation machinery, such as Ubc-9, have been shown to be overexpressed in ovarian, prostate and colon cancers (Eifler and Vertegall 2015). This further increases the attractiveness of SUMO components as cancer drug targets.
Progress has recently been made in applying SUMOylation inhibitors for clinical purposes; Bernstock et al. (2017) recently demonstrated the potential use of the global SUMOylation inhibitor Topotecan as an adjuvant therapy for certain types of glioblastoma multiforme (GBM). This study only provides a preview of the potential of SUMO inhibitors as therapeutic agents but highlights their potential for treating cancer.
Studying Cell Signaling Pathways?
To assist with your cell signaling research we offer many products including a comprehensive range of anti-EGFR antibodies and resources, such as protocols, webinars, tips and tricks, for key cancer applications, such as immunohistochemistry, immunoprecipitation and western blotting.
References
Bernstock JD et al. (2017). Topotecan is a potent inhibitor of SUMOylation in glioblastoma multiforme and alters both cellular replication and metabolic programming. Sci Rep 7, 7425.
Bouchard C et al. (1998). Control of cell proliferation by Myc. Trends Cell Biol 8, 202-206.
Cremona CA et al. (2012). Sumoylation and the DNA Damage Response. Biomolecules 2, 376-388.
Eifler K and Vertegall ACO (2015). SUMOylation-mediated regulation of cell cycle progression and cancer. Trends Biochem Sci 40, 779-793.
González-Prieto R et al. (2015). c-Myc is targeted to the proteasome for degradation in a SUMOylation-dependent manner, regulated by PIAS1, SENP7 and RNF4. Cell Cycle 14, 1859-72.
Hendriks IA and Vertegaal ACO (2016). A comprehensive compilation of SUMO proteomics. Nat Rev Mol Cell Biol 17, 581-95.
Huang J et al. (2012). SUMO1 modification of PTEN regulates tumorigenesis by controlling its association with the plasma membrane. Nat Commun 3, 911.
Leslie NR and Longy M (2016). Inherited PTEN mutations and the prediction of phenotype. Semin Cell Dev Biol Vol 52, 30-38.
Li R et al. (2013). Akt SUMOylation regulates cell proliferation and tumorigenesis. Cancer Res 73, 5742-53.
Miller DM et al. (2012). c-Myc and Cancer Metabolism. Clin Cancer Res 18:5546-5553.
Myatt SS et al. (2014). SUMOylation inhibits FOXM1 activity and delays mitotic transition. Oncogene 33, 4316-4329.
Nandi D et al. (2017). FoxM1: Repurposing an oncogene as a biomarker. Semin Cancer Biol pii: S1044-579X(17)30061-5.
Schimmel J et al. (2014). Uncovering SUMOylation dynamics during cell-cycle progression reveals FoxM1 as a key mitotic SUMO target protein. Mol Cell 53, 1053-66.
Song MS et al. (2012). The functions and regulation of the PTEN tumour suppressor. Nature Rev Mol Cell Biol 13, 283-296.
Yang C et al. (2013). Inhibition of FOXM1 transcription factor suppresses cell proliferation and tumor growth of breast cancer. Cancer Gene Ther 20, 117-124.
Zona S et al. (2014). FOXM1: An emerging master regulator of DNA damage response and genotoxic agent resistance. Biochim Biophys Acta 1839:1316-1322.
You may also be interested in...

View more Cell Cycle or Tips&Tricks blogs















