
Popular topics

-
References
Fensterl VS et al. (2015). No love lost between viruses and interferons. Annu Rev Virol 2, 549-72.
Glanz A et al. (2020). High throughput screening of FDA-approved drug library reveals the compounds that promote IRF3-mediated pro-apoptotic pathway inhibit virus replication. Viruses 12, 442.
Hemmi H et al. (2002). Small anti-viral compounds activate immune cells via the TLR7 MyD88-dependent signaling pathway. Nat Immunol 3, 196-200.
Hornung V and Latz E (2010). Intracellular DNA recognition. Nat Rev Immunol 10, 123-30.
Majumdar T et al. (2015). Induction of interferon-stimulated genes by IRF3 promotes replication of Toxoplasma gondii. PLoS Pathog 1, e1004779.
Schoggins JW and Rice CM (2011). Interferon-stimulated genes and their antiviral effector functions. Current Opinion in Virology 1, 519-525.
Subramanian G et al. (2018). A new mechanism of interferon's antiviral action:Induction of autophagy, essential for paramyxovirus replication, is inhibited by the interferon stimulated gene, TDRD7. PLoS Pathog 14, e1006877
Wu J and Chen ZJ (2014). Innate immune sensing and signaling of cytosolic nucleic acids. Annu Rev Immunol 32, 461-88.
Zheng C (2018). Evasion of cytosolic DNA-stimulated innate immune responses by herpes simplex virus 1. J Virol 92, e00099-17.
How Do Pattern Recognition Receptors Help Mount an Immune Response?
The fear of the unknown is quite common and knowing that almost every cell in your human body can fight back against a viral attack is a great reassurance. For each of these cells to do so, recognition of the invader is an important step. This blog by guest contributor Gayatri Subramanian, describes how the host innate immune system tackles viral pathogens through recognition of the virus.
What Do the Cellular Sensors Look Like?
Just like in a chess line up where the most valuable pieces are protected by the pawns (and make the first move) pattern recognition receptors (PRRs) act as a first line of defense in the innate immune system, before the onset of interferon and inflammatory response. These receptors are activated during viral infection when they recognize and bind a trigger, or a pathogen associated molecular pattern (PAMP) — a molecular signature, or viral DNA, or RNA.
What Is Recognized by the PRRs?
PRRs as their name suggests, recognize certain patterns within foreign genomic material. This material can be long dsRNA, short dsRNA, ssRNA with either a 5’ phosphorylation or a 3’ phosphorylation, dsDNA, ssDNA, oxidized DNA, ssDNA as well as a DNA-RNA hybrid. Characteristics specific to these nonself, genetic materials include the length of the genomic material, the presence of a cap, or a secondary structure (Hornung and Latz 2010).
RNA sensing in the cytosol is carried out by RIG-I like receptors (RLRs) and there are three members in this family – the most common ones are RIG-I, MDA5, and LGP2. RIG-I can recognize unmodified 5’ phosphorylated RNA common to negative sense RNA viruses, while MDA5 is reported in the recognition of dsRNA seen in case of positive sense RNA virus infection. Other RNA sensors include Protein Kinase R (PKR), 2’5’-oligoadenylate synthase (OAS), and RNaseL.
Toll-like receptors (TLRs) are present on the endosomes and are listed as TLR1 through 9 – each recognizing a different foreign component. For example, TLR3 detects dsRNA and TLR7/8 recognize ssRNA.
DNA sensors include cytosolic sensors cGAS/STING, AIM2 as well as helicase sensors such as DHX36, DDX41 and DDX60. A nucleic sensor of nonself-DNA is IFI16, while on the endosome TLR9 recognizes DNA as well as DNA-RNA hybrids. The expression of these PRRs is characteristic of specific cell types, where the TLRs are primarily seen on dendritic cells and the cytosolic DNA and RNA sensors are more ubiquitously expressed in other cell types. In these cells, some PRRs are dispensable due to the redundancy in function by some others (Fensterl et al. 2015, Wu and Chen 2014).
What Happens After Recognition?
After the binding step, several conformational changes occur that result in the activation of a downstream signaling cascade that eventually results in the infected cell mounting an antiviral response (Figure 1). This signaling cascade includes adaptor proteins and transcription factors downstream of PPR activation. The activation of these receptors and the downstream adaptor proteins, as well as transcription factors, occurs through post-translational modifications like phosphorylation, dephosphorylation, and ubiquitination. (Majumdar et al. 2015).
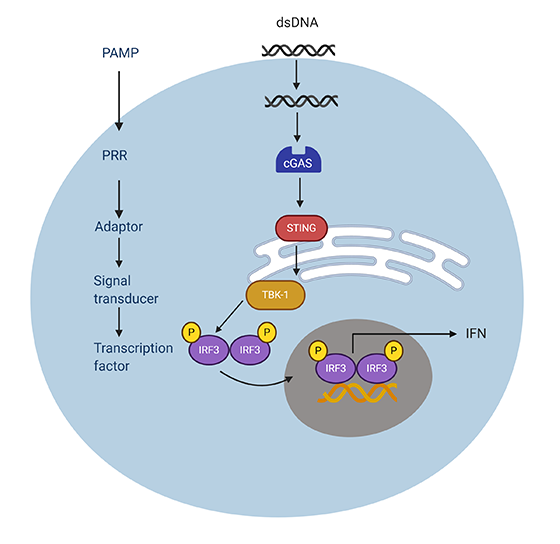
Fig.1. An example of how PRR leads to an immune response. A PAMP, in this case dsDNA, produced by the pathogen is recognized by the PRR – cGAS, which activates STING. STING (the adaptor) forms a complex with TBK-1 (signal transducer) which then phosphorylates and activates IRF3 (transcription factor). IRF3 then translocates to the nucleus to induce the transcription of IFN.
Can Viruses Escape These Consequences?
Viruses have evolved evasion techniques that help avoid recognition by these PRRs. This is possible by the masking of genetic material or by the degradation of proteins within the recognition pathway. Herpes Simplex Virus – 1 (HSV-1) is well known for its evasion of the innate immune response. This virus is a DNA virus and the PRRs that help recognize it include DNA sensors, such as STING and IFI16, and as the DNA virus replication involves an RNA step (in transcription and translation of viral proteins), also RNA sensors like PKR. In spite of this broad range of cellular sensors in HSV-1 recognition, this virus effectively escapes the host response by means of its proteins such as vhs (virus host shutoff), ICP0, ICP27, ICP34.5, Us11 and UL36. This type of antagonism results in decreased IFN production and conditions conducive for viral replication (Zheng 2018).
Can We Improve Immune Response by Enhancing Recognition?
In the context of a viral infection, the innate immune response functions immediately through the identification of nonself and elicite a broad response that helps mitigate the incoming pathogen. As a therapeutic approach, aiding the recognition, enhancing the response of recognition pathways, and preventing the antagonism by these pathogens, are a few possible directions. A study by Hemmi et al. (2002) identified ligands of TLR7 – imiquimod and R-848 – belonging to the family of imidazoquinoline that resulted in interferon and cytokine production, resulting in antiviral activity. Recently, Glanz et al. (2020) carried out a screen to identify small molecule enhancers of a RIG-I dependent apoptotic pathway that increased cellular apoptosis to reduce virus replication. The molecular mechanism of the drug doxorubicin is to target downstream of pathogen recognition, at the transcription factor stage. Thus, understanding signaling of viral recognition pathways can serve as another approach in antiviral drug design.
Want to Know More about TLRs?
Toll-like receptors are microbial-sensing proteins that recognize a wide range of pathogens, they are a critical protein in the initial protection against infection. Read our mini-review to learn about this family of proteins.
References
Fensterl VS et al. (2015). No love lost between viruses and interferons. Annu Rev Virol 2, 549-72.
Glanz A et al. (2020). High throughput screening of FDA-approved drug library reveals the compounds that promote IRF3-mediated pro-apoptotic pathway inhibit virus replication. Viruses 12, 442.
Hemmi H et al. (2002). Small anti-viral compounds activate immune cells via the TLR7 MyD88-dependent signaling pathway. Nat Immunol 3, 196-200.
Hornung V and Latz E (2010). Intracellular DNA recognition. Nat Rev Immunol 10, 123-30.
Majumdar T et al. (2015). Induction of interferon-stimulated genes by IRF3 promotes replication of Toxoplasma gondii. PLoS Pathog 1, e1004779.
Schoggins JW and Rice CM (2011). Interferon-stimulated genes and their antiviral effector functions. Current Opinion in Virology 1, 519-525.
Subramanian G et al. (2018). A new mechanism of interferon's antiviral action:Induction of autophagy, essential for paramyxovirus replication, is inhibited by the interferon stimulated gene, TDRD7. PLoS Pathog 14, e1006877
Wu J and Chen ZJ (2014). Innate immune sensing and signaling of cytosolic nucleic acids. Annu Rev Immunol 32, 461-88.
Zheng C (2018). Evasion of cytosolic DNA-stimulated innate immune responses by herpes simplex virus 1. J Virol 92, e00099-17.
You may also be interested in...

View more Immunology or Guest Blog blogs















