
Popular topics

-
References
Ando R et al. (2004). Regulated fast nucleocytoplasmic shuttling observed by reversibly protein highlighting. Science 306, 1370-1373.
Ballestrem C et al. (1998). Actin dynamics in living mammalian cells. J Cell Sci 111 (Pt2), 1649-1658.
Chalfie M et al. (1994). Green fluorescent protein as a marker for gene expression. Science 263, 802-805.
Chalfie M and Kain S (2006). Green fluorescent protein: Properties, applications, and protocols (New Jersey: Wiley-Interscience).
Chudakov DM et al. (2003). Kindling fluorescent proteins for precise in vivo photolabeling. Nat Biotechnol 21, 191-194.
Davenport D and Nicol JAC (1955). Luminescence in hydromedusae. Proc R Soc Lond B 144, 399-411.
Hoffman RM (2005). The multiple uses of fluorescent proteins to visualize cancer in vivo. Nat Rev Cancer 5, 796-806.
Lippincott-Schwartz J and Patterson GH (2003). Development and use of fluorescent protein markers in living cells. Science 300, 87-91.
Livet J et al. (2007). Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature 450, 56-62.
Lukyanov KA et al. (2005). Innovation: Photoactivatable fluorescent proteins. Nat Rev Mol Cell Biol 6, 885-891.
Matz MV et al. (1999). Fluorescent proteins from nonbioluminescent Anthozoa species. Nat Biotechnol 17, 969-973.
Morin JG and Hastings JW (1971). Energy transfer in a bioluminescent system. J Cell Physiol 77, 313-318.
Prasher DC et al. (1992). Primary structure of the Aequorea victoria green-fluorescent protein. Gene 111, 229-233.
Rizzuto R et al. (1995). Chimeric green fluorescent protein as a tool for visualizing subcellular organelles in living cells. Curr Biol 5, 635-642.
Royal Swedish Academy of Sciences (2008). How the jellyfish's green light revolutionised bioscience.https://www.nobelprize.org/uploads/2018/06/popular-chemistryprize2008.pdf, accessed December 1, 2016.
Sekar RB and Periasamy A (2003). Fluorescence resonance energy transfer (FRET) microscopy imaging of live cell protein localizations. J Cell Biol 160, 629-633.
Shcherbo D et al. (2007). Bright far-red fluorescent protein for whole body imaging. Nat Methods 4, 741-746.
Shimomura O et al. (1962). Extraction, purification and properties of aequorin, a bioluminescent protein from the luminous hydromedusan, Aequorea. J Cell Comp Physiol 59, 223-229.
Tsien RY (1998). The green fluorescent protein. Annu Rev Biochem 67, 509-544.
Illuminating biomedical research — the story of fluorescent proteins
Aequorea victoria is a bioluminescent hydrozoan jellyfish found in the waters off the west coast of North America. Its brilliantly luminous nature signifies how it has revolutionized biomedical research.
In 1955, Davenport and Nicol were the first to report the presence of a fluorescent component in the bioluminescent tissues of A. victoria (Davenport and Nicol 1955). Around 5 years later, on his first day as a research assistant in the lab of Dr. Frank Johnson, Dr. Osamu Shimomura was tasked with studying the bioluminescence of this jellyfish. This led to the isolation of the bioluminescent protein aequorin and the now famous green fluorescent protein (GFP) (Shimomura et al. 1962).
Later studies demonstrated that aequorin emits blue light in the presence of calcium, and this light is then transduced to green fluorescence by the aptly named GFP (Chalfie and Kain 2006). After the discovery of GFP from A. victoria, other GFPs were isolated from the hydrozoan jellyfish Obelia and the sea pansy Renilla reniformis (Morin and Hastings 1971).
As the pioneer fluorescent proteins, GFPs were quickly adapted in biomedical research labs worldwide. Some of the biggest research milestones that paved the way for GFPs as research tools include the cloning and sequencing of the cDNA of A. victoria GFP as well as the demonstration that this GFP could be functionally expressed in other biological organisms and used as a marker of gene expression (Chalfie et al. 1994, Prasher et al. 1992).
By the mid to late 1990s, the ubiquitous use of GFP as a fluorescent protein tag for localization studies highlighted a need for different color emitting fluorescent proteins that would enable dual labeling experiments and co-localization studies.
Primarily through the prominent work of the late Dr. Roger Tsien, a series of A. victoria GFP variants with distinct excitation and emission maxima were developed. Some of these variants include a cyan excitable enhanced GFP (EGFP), blue fluorescent protein (BFP), cyan fluorescent protein (CFP), yellow fluorescent protein (YFP) and a violet-excitable variant called Sapphire (Tsien 1998). These GFP variants were engineered to start fluorescing faster and to be brighter than wild type GFP.
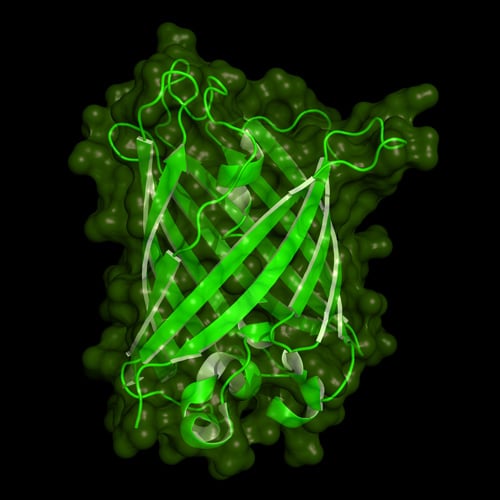
Cartoon model of the protein structure of green fluorescent protein
The popularity of these GFP proteins also led to the pursuit of GFP-like fluorescent proteins in other organisms. In 1999, a team of researchers led by Dr. Sergey Lukyanov at the Russian Academy of Science demonstrated that non-luminescent reef Anthozoa contain fluorescent proteins in colors ranging from cyan to red (Matz et al. 1999). DsRed from Discosoma sp. mushroom was the most popular red fluorescent protein isolated from Anthozoa (Matz et al. 1999).
In 2007, the same lab identified another red fluorescent protein that is significantly brighter than DsRed called Katushka (Shcherbo et al. 2007). The monomeric form of this tetramer protein is called mKate. Dr. Roger Tsien later generated derivatives of DsRed to create a fruit series of fluorescent proteins, which include mCherry, mPlum, mStrawberry, mOrange and mCitrine.
How fluorescent proteins have advanced scientific discovery
The most common, and now routine, application of fluorescent proteins by scientists involves their use in microscopy for imaging the localization and dynamics of specific organelles or as recombinant proteins in live cells (Ballestrem et al. 1998, Rizzuto et al. 1995). Standard molecular biology techniques are applied for the visualization of proteins of interest using fluorescent proteins.
Essentially, the gene encoding a fluorescent protein is fused to the cDNA of a specific target protein or peptide that localizes to a particular tissue. A plasmid containing this chimeric gene is then transfected into mammalian cells to produce the corresponding chimeric protein.
This protein localizes to the target organelle and thus makes it fluorescent. This technique has provided researchers with new opportunities to study various biological processes such as development in a number of organisms.
The availability of a wide range of fluorescent proteins has also led to novel technologies. For instance, by applying the phenomenon of fluorescence resonance energy transfer (FRET), scientists have developed methods using fluorescent proteins to determine whether two proteins are located in a distance of 10 nm of each other (Sekar and Periasamy 2003).
Another technique known as FLiP (fluorescence loss in photobleaching) utilizes a fluorescent protein found in corals called Dronpa, and allows researchers to activate and monitor protein activity (Lippincott-Schwartz and Patterson 2003).
Fluorescent proteins called "optical highlighter proteins" that change their emission upon irradiation have been applied for imaging sub-populations of cells during development as well as imaging of fast protein dynamics (Lukyanov et al. 2005, Ando et al. 2004, Chudakov et al. 2003).
Scientists have also generated transgenic animals expressing visible amounts of fluorescent proteins. Perhaps the most famous example of such a model is the "Brainbow" mouse, in which each cell in the brain of a living mouse was colored using fluorescent proteins (Livet et al. 2007). Fluorescent transgenic animals have been used to understand various biological processes in normal and diseased cells. Particularly, fluorescent mice have been especially useful in cancer research (Hoffman 2005).
These examples demonstrate that fluorescent proteins have expanded the capabilities of scientists, thus allowing observations that would not have been possible without them.
The important contribution of fluorescent proteins to advancing science was recognized with the award of the 2008 Nobel Prize in Chemistry for "the discovery and development of green fluorescent protein, GFP" to Osamu Shimomura, Martin Chalfie and Roger Tsien.
GFP has been referred to as "a guiding star for biochemists, biologists, medical scientists and other researchers," and for this science is indebted to the A. Victoria jellyfish in which it was discovered over 50 years ago (Royal Swedish Academy of Sciences 2008).
Find fluorescent proteins for your research
To support scientists using fluorescent proteins in their research, Bio-Rad now offers antibodies to common fluorescent proteins including BFP, CFP, DsRed, GFP and YFP. These antibodies can be used to determine the expression levels of different fluorescent proteins by western blotting.
References
Ando R et al. (2004). Regulated fast nucleocytoplasmic shuttling observed by reversibly protein highlighting. Science 306, 1370-1373.
Ballestrem C et al. (1998). Actin dynamics in living mammalian cells. J Cell Sci 111 (Pt2), 1649-1658.
Chalfie M et al. (1994). Green fluorescent protein as a marker for gene expression. Science 263, 802-805.
Chalfie M and Kain S (2006). Green fluorescent protein: Properties, applications, and protocols (New Jersey: Wiley-Interscience).
Chudakov DM et al. (2003). Kindling fluorescent proteins for precise in vivo photolabeling. Nat Biotechnol 21, 191-194.
Davenport D and Nicol JAC (1955). Luminescence in hydromedusae. Proc R Soc Lond B 144, 399-411.
Hoffman RM (2005). The multiple uses of fluorescent proteins to visualize cancer in vivo. Nat Rev Cancer 5, 796-806.
Lippincott-Schwartz J and Patterson GH (2003). Development and use of fluorescent protein markers in living cells. Science 300, 87-91.
Livet J et al. (2007). Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature 450, 56-62.
Lukyanov KA et al. (2005). Innovation: Photoactivatable fluorescent proteins. Nat Rev Mol Cell Biol 6, 885-891.
Matz MV et al. (1999). Fluorescent proteins from nonbioluminescent Anthozoa species. Nat Biotechnol 17, 969-973.
Morin JG and Hastings JW (1971). Energy transfer in a bioluminescent system. J Cell Physiol 77, 313-318.
Prasher DC et al. (1992). Primary structure of the Aequorea victoria green-fluorescent protein. Gene 111, 229-233.
Rizzuto R et al. (1995). Chimeric green fluorescent protein as a tool for visualizing subcellular organelles in living cells. Curr Biol 5, 635-642.
Royal Swedish Academy of Sciences (2008). How the jellyfish's green light revolutionised bioscience.https://www.nobelprize.org/uploads/2018/06/popular-chemistryprize2008.pdf, accessed December 1, 2016.
Sekar RB and Periasamy A (2003). Fluorescence resonance energy transfer (FRET) microscopy imaging of live cell protein localizations. J Cell Biol 160, 629-633.
Shcherbo D et al. (2007). Bright far-red fluorescent protein for whole body imaging. Nat Methods 4, 741-746.
Shimomura O et al. (1962). Extraction, purification and properties of aequorin, a bioluminescent protein from the luminous hydromedusan, Aequorea. J Cell Comp Physiol 59, 223-229.
Tsien RY (1998). The green fluorescent protein. Annu Rev Biochem 67, 509-544.
You may also be interested in...

View more Applications or Immunofluorescence blogs















