
Popular topics

How to Pull-Down Your Target Protein and Interactors Successfully

Immunoprecipitation (IP), also known as pull-down, is the process of precipitating a protein antigen out of solution, usually a whole cell lysate. This blog will take you through the basics of IP so you can get successful results from the outset.
The first step is to select an antibody targeting your protein of interest and couple it to beads coated with either Protein A or Protein G. The antibody coupled to the beads binds to the protein of interest in your sample lysate and can be used to isolate your target. The lysate is incubated with the beads and any unbound proteins are removed through a series of wash steps. Finally, the target protein and any interaction partners that may be bound to it are eluted off the beads (Figure1).
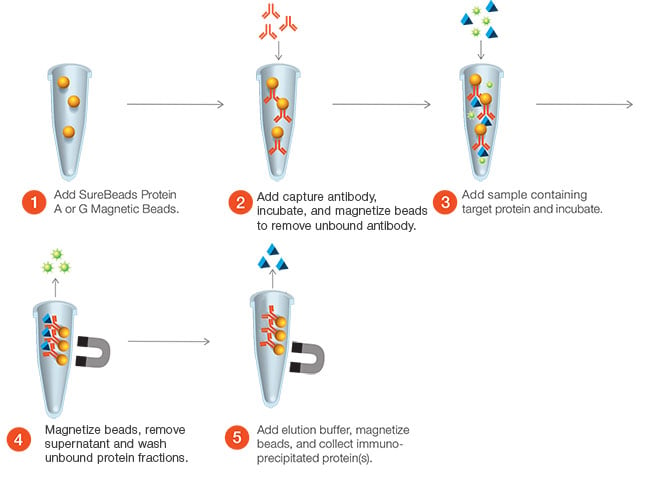
Fig. 1. Example of a method for performing IP using Bio-Rad’s SureBeads Magnetic Beads.
Co-immunoprecipitation (co-IP) uses antibodies to indirectly capture any proteins that are bound to the target protein. Co-IP is commonly used to identify new interaction partners and characterize protein complexes. IPs may also be used to identify post-translational modifications or increase the concentration of a particular protein in a sample. Whatever the aim of your IP experiment, the following key steps are critical to the successful pull-down of your target protein.
1. Lysate Preparation
The ideal lysis buffer should conserve the native conformation of your protein of interest while also efficiently lyzing your cells. It is crucial to consider the nature of your protein of interest when choosing a lysis buffer. There are two main types of lysis buffer: denaturing and non-denaturing.
Non-denaturing lysis buffers contain relatively gentle detergents such as NP-40 or Triton X-100 and are suitable if your target protein is soluble in these detergents. This type of buffer tends to maintain the native conformation of the target protein. However, there are exceptions like some certain cytoskeletal proteins, which may not be solubilized.
Denaturing lysis buffers, such as RIPA buffer, contain components like SDS. These buffers are harsher and can denature some proteins. However, they are ideal for the IP of nuclear proteins as disruption of the nuclear membrane is required to gain access to these proteins.
You should protect your protein from being proteolytically degraded by adding protease inhibitors to your lysis buffer. If you are intending to analyze phosphorylated proteins, make sure that you maintain the protein’s modification status by adding phosphatase inhibitors to prevent dephosphorylation of your target.
The concentration of your prepared lysate may also influence the IP efficiency. Generally, a higher lysate concentration results in more of your protein of interest being present in your final eluted IP sample. In order to improve the detection of rare events like phosphorylation, or weakly expressed proteins, you can reduce the amount of lysis buffer you add to your cells. However, if your protein of interest is highly abundant you may saturate your IP antibody at relatively low lysate concentrations. Increasing the lysate concentration will not increase IP efficiency.
2. Antibody Selection
As with any experiment, antibody selection is crucial for success. If available, you should choose an antibody that has been validated for use in IP. However, depending on your target, it may be difficult to find an IP tested antibody. If you cannot use an IP validated antibody, a good alternative is to choose an antibody that has been shown to work in another application that requires detection of the native protein conformation, like flow cytometry.
3. Selecting the Best Bead Type
Several types of beads are available for performing IP. Each bead type has different advantages and disadvantages.
Magnetic beads are more expensive than agarose or sepharose beads but, as less antibody may be required to get results, so you could save on antibody costs. The risk of losing beads during wash steps is also reduced as the beads are held in place by a magnet, making it significantly easier than pelleting agarose beads by centrifugation and manually removing the supernatant. IPs performed with magnetic beads generally result in reduced levels of non-specific binding and therefore can aid in the identification of novel protein-protein interactions.
Check that the antibody isotype you are using is compatible with your beads as not all isotypes bind well to both Protein A and Protein G. While some isotypes, such as mouse IgG2a bind equally well to both Protein A and Protein G, enabling you to use beads coated with either, other isotypes such as rat IgG2a only demonstrate binding affinity to either Protein A or Protein G. It is then critical to use beads coated with the correct protein.
4. Elution Method
The most common types of elution buffers used for IP experiments contain SDS or are glycine-based. When you are unsure of the most suitable elution method for your protein, SDS-based elution buffers provide a good starting point as they are highly efficient. However, they can result in higher background in subsequent western blot detection than other buffer types as they often elute the IP antibody from beads as well as the bound proteins, resulting in the presence of antibodies in your IP sample.
As SDS-based buffers are denaturing, they are not recommended if you require your protein in the native conformation. Elution steps require heat treatment of the sample, which can disrupt the bonds, therefore rendering this method unsuitable for studying some protein-protein interactions.
If you do want to use downstream applications that rely on the native conformation of your protein of interest, glycine-based elution buffers are a better choice. They are non-denaturing and rely on the pH change to gently dissociate bound proteins from the beads. These buffers also tend to result in lower background in subsequent western blot detection, as the IP antibody concentration in the final sample is lower compared to SDS-based buffers. Take care though as this buffer type is not universally effective and may change the conformation of some proteins.
Looking for IP Protocols and Resources?
I hope that by considering these main areas of an IP experiment carefully you will be able to perform your experiments successfully. In addition. Bio-Rad has detailed protocols, tips, and troubleshooting advice on our dedicated immunoprecipitation application resources page.
View the IP Resources
You may also be interested in...

View more Applications or Tips&Tricks blogs















