
Popular topics

-
References
Burgess SM and Powers T (2017). Budding Yeast Saccharomyces Cerevisiae as a Model Genetic Organism. eLS 1-12.
Coelho-Silva L et al. (2017). SUMOylation and calcium signaling: potential roles in the brain and beyond. Neuronal Signaling Vol 1, issue 3.
Da Silva-Ferrada E et al. (2013). Analysis of SUMOylated proteins using SUMO-traps. Scientific Reports 3, Article number 1690.
Gardner RD et al. (2001). The spindle checkpoint of the yeast Saccharomyces cerevisiae requires kinetochore function and maps to the CBF3 domain. Genetics 157, 1493-502.
Gareau JR and Lima CD (2010). The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat Rev Mol Cell Biol 11, 861-871.
Hendriks IA and Vertegaal ACO (2015). SUMO in the DNA damage response. Oncotarget 6, 15734-15735.
Joseph J et al. (2002). SUMO-1 targets RanGAP1 to kinetochore and mitotic spindles. J Cell Biol 156, 595-602.
Li SJ and Hochstrasser M (1999). A new protease required for cell-cycle progression in yeast. Nature 398, 246-251.
Mahajan R et al. (1997). A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell 88, 97-107.
Matunis MJ et al. (1996). A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J Cell Biol 135, 1457-1470.
Meluh PB and Koshland D (1995). Evidence that the MIF2 gene of Saccharomyces cerevisiae encodes a centromere protein with homology to the mammalian centromere protein CENP-C. Mol Biol Cell 6, 793-807.
Nacerddine K et al. (2005). The SUMO pathway is essential for nuclear integrity and chromosome segregation in mice. Dev Cell 9, 769-779.
Seufert W et al. (1995). Role of a ubiquitin-conjugating enzyme in degradation of S- and M-phase cyclins. Nature 373, 78-81.
Xiao Y et al. (2015). Can your protein be sumoylated? A quick summary and important tips to study SUMO-modified proteins. Anal Biochem 15, 95-97.
SUMOylation: a Heavyweight in the Regulation of Cell Cycle Progression and Cell Proliferation – Part One

Studying SUMOylation?
Check out our podcast on SUMOylation, including our tips to help you study this post translational modification. Spend your experimental break wisely and get some science on the go in less than 5 minutes.
Duration: 04:03 | File size: 3581KB | Recommended browsers: Google Chrome and Mozilla Firefox.
In the first part of our two-part series on SUMOylation we discuss the discovery of SUMO and the importance of SUMOylation in cell cycle regulation.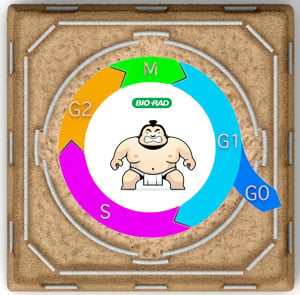
SUMO (small ubiquitin-related modifier) proteins are members of a family of structurally related protein modifiers, which also includes ubiquitin and NEDD8.
Currently five SUMO proteins have been identified; these are SUMO-1, SUMO-2, SUMO-3, SUMO-4 and SUMO-5 (Coelho-Silva et al. 2017). Conjugation of these SUMO proteins to lysine (K) residues present on protein substrates results in various functional changes. For example, SUMOylation has been reported to alter the subcellular localization and protein interactions of its substrates (Gareau and Lima 2010).
SUMOylation is a rapid and reversible modification and has a well-documented role in transcriptional regulation. SUMOylation is therefore an effective response tool to environmental and cellular stresses, such as DNA damage (Hendriks and Vertegaal 2015). Moreover, SUMO has been linked with cell cycle progression right from its initial discovery.
From Yeast to Mammals – Importance of SUMOylation in Cell Cycle Regulation
The budding yeast, Saccharomyces cerevisiae, has been a popular model organism for over 50 years (Burgess and Powers 2017). Discoveries made in yeast have majorly advanced cell biology research and have significantly contributed to our current understanding of the eukaryotic cell cycle. It is perhaps not surprising that the first SUMO homolog, SMT3, was discovered in S. cerevisiae. SMT3 was initially identified in a genetic suppressor screen of mutations in the MIF2 gene, which encodes a centromere protein essential for cell cycle progression (homolog of the mammalian CENP-C protein) (Gardner et al. 2001, Meluh and Koshland 1995).
The experiment in which SMT3 was identified implied a role for the protein in cell cycle progression and regulation. This theory gained traction when the S. cerevisiae SUMO-conjugating enzyme, Ubc9, was reported by Seufert et al. (1995) to play a role in the “degradation of S- and M-phase cyclins "and the progression through mitosis. Further evidence came from Li and Hochstrasser (1999), who showed that the ubiquitin like specific protease, ULP1, was essential for the G2 to M phase transition of the cell cycle.
It was not long after these initial experiments in yeast that SUMOylated proteins were discovered in mammals. The first one to be identified was the Ran GTPase-activating protein 1, RanGAP1 (Matunis et al. 1996). Western blotting and imaging experiments showed that unmodified 70 kDa RanGAP1 was exclusively localized in the cytoplasm, while the modified 90 kDa variant co-localized to the “cytoplasmic fibers of the nuclear pore complex” (Matunis et al. 1996). The moiety conjugated to RanGAP1 was subsequently named SUMO-1 by Frauke Melchior’s group (Mahajan et al. 1997) and shown to be critical for localizing RanGAP1 to mitotic spindles and kinetochores (Joseph et al. 2002).
Further evidence for the essential role of SUMOylation in mammals came from a study conducted by Nacerddine et al. (2005), which showed that Ubc-9 deficient mice ”die at the postimplantation stage”. Although cultured Ubc-9 deficient blastocytes were viable, these cells showed severe mitotic defects, such as impaired chromosome segregation (Nacerddine et al. 2005). Furthermore, Ubc-9 deficient cells showed a RanGAP1 distribution defect as the protein no longer localized at the nuclear pore complex (Nacerddine et al. 2005). Thus, confirming the earlier result by Mahajan et al. (1997) that SUMOylation is required for localization of RanGAP1 to this region.
Over the next decade, the importance of SUMOylation in orchestrating cell cycle progression has been continuously demonstrated and many key mediators have been subsequently shown to be regulated through this modification. In our next blog post we discuss how SUMOylation regulates key cell signaling proteins and how their aberrant signaling can lead to the development of cancer.
Tips and Tricks for Studying SUMOylation
- Sample preparation: to preserve SUMOylated proteins, inhibit the effects of deSUMOylation enzymes (SENPs) by adding the isopeptidase inhibitors, N-ethylamaleimide (NEM) and/or 2-iodoacetamide (IAA), to lysis buffers (Xiao et. 2015, Da Silva-Ferrada et al. 2013). Caution should be taken as these inhibitors may also inhibit deubiquitinating (DUBs) enzymes
- Western blot detection: as only a small percentage of intracellular proteins are SUMOylated at any time, we recommend enrichment of your SUMOylated protein(s) of interest by performing an immunoprecipitation (IP) experiment prior to western blot detection. For best IP results use highly concentrated cell lysates (e.g. 5 x 106 – 1 x 107 cells in 300 µl of lysis buffer) (Xiao et al. 2015). Additional information on IP procedures can be found on the IP application page or in our webinar. We also have collated some of our most popular IP tips in the IP tips and tricks document
- Assay selection: detection of immunoprecipitated proteins with the help of SUMO antibodies may provide an initial indication as to whether a protein is SUMOylated or not. However as with many experimental assays, there are limitations to this approach. To complement this assay, SUMO-traps/SUMO binding entities (SUBEs) can be used to capture endogenous SUMOylated proteins (Da Silva-Ferrada et al. 2013). SUMO-traps contain SUMO interacting motifs (SIMs), which have been multiplied and conjugated to the GST carrier protein, enabling the specific pull-down of SUMOylated proteins (Da Silva-Ferrada et al. 2013)
Studying cell proliferation?
We offer many products including Mouse Anti-BrdU Antibody, clone Bu20a, and Rabbit Anti-BrdU Antibody (AHP2405) for cell proliferation analysis. We have a range of comprehensive cell proliferation resources, which include protocols, blog posts and our popular podcast, to assist your cell proliferation experiments.
SUMOylation Blog – Part Two
References
Burgess SM and Powers T (2017). Budding Yeast Saccharomyces Cerevisiae as a Model Genetic Organism. eLS 1-12.
Coelho-Silva L et al. (2017). SUMOylation and calcium signaling: potential roles in the brain and beyond. Neuronal Signaling Vol 1, issue 3.
Da Silva-Ferrada E et al. (2013). Analysis of SUMOylated proteins using SUMO-traps. Scientific Reports 3, Article number 1690.
Gardner RD et al. (2001). The spindle checkpoint of the yeast Saccharomyces cerevisiae requires kinetochore function and maps to the CBF3 domain. Genetics 157, 1493-502.
Gareau JR and Lima CD (2010). The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat Rev Mol Cell Biol 11, 861-871.
Hendriks IA and Vertegaal ACO (2015). SUMO in the DNA damage response. Oncotarget 6, 15734-15735.
Joseph J et al. (2002). SUMO-1 targets RanGAP1 to kinetochore and mitotic spindles. J Cell Biol 156, 595-602.
Li SJ and Hochstrasser M (1999). A new protease required for cell-cycle progression in yeast. Nature 398, 246-251.
Mahajan R et al. (1997). A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell 88, 97-107.
Matunis MJ et al. (1996). A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J Cell Biol 135, 1457-1470.
Meluh PB and Koshland D (1995). Evidence that the MIF2 gene of Saccharomyces cerevisiae encodes a centromere protein with homology to the mammalian centromere protein CENP-C. Mol Biol Cell 6, 793-807.
Nacerddine K et al. (2005). The SUMO pathway is essential for nuclear integrity and chromosome segregation in mice. Dev Cell 9, 769-779.
Seufert W et al. (1995). Role of a ubiquitin-conjugating enzyme in degradation of S- and M-phase cyclins. Nature 373, 78-81.
Xiao Y et al. (2015). Can your protein be sumoylated? A quick summary and important tips to study SUMO-modified proteins. Anal Biochem 15, 95-97.
You may also be interested in...

View more Cell Cycle or Tips&Tricks blogs















