Article: Biomarker Assay Development Using Highly Specific Recombinant Antibodies
-
Monoclonal Generation
-
Custom Recombinant Monoclonal Antibody Generation
-
Webinars, Videos and Technical Articles
- Webinar: Overcome the Challenges of PK Assay Development Using TrailBlazer Antibodies
- Webinar: Generation of SARS-CoV-2 antibodies in multiple formats within four weeks
- Webinar: Recombinant Antibodies with SpyTag Technology
- Webinar: Transform bioanalytical assays with TrailBlazer Antibodies
- Webinar: Control your critical antibody reagents and avoid assay failure
- Webinar: Improve your antibody drug development assays
- Webinar: Optimize your assays using recombinant antibodies selected for desired affinity
- Webinar: The making of recombinant anti-idiotypic antibodies for high performance in bioanalytical assays
- Webinar: Human recombinant antibodies as positive controls and calibrators
- Webinar: How to overcome assay challenges using custom recombinant antibodies
- Webinar: Generation of high affinity recombinant antibodies for application in immuno-MRM
- Video: Generating anti-idiotypic antibodies for bioanalytical assays
- Video: Best practices for characterization and QC of anti-idiotypic antibodies for bioanalysis
- Video: Generation of drug-target complex specific antibodies
- Video: Antibodies for CAR-T cell therapy development
- Article: Monitoring antibody immune responses against biotherapeutic drugs
- Article: Effective tools for drug monitoring assays
- Article: An accelerated approach to sensitive ADA assays
- Article: Isolation of enzyme active site-specific recombinant antibodies by guided selection
- Article: Biomarker Assay Development using Highly Specific Recombinant Antibodies
- Article: Recombinant antibodies as standards for immunodiagnostic assays
- Article: High affinity antibodies for peptide enrichment immuno-MRM
- Article: Generation of antibodies against self-antigens
- Article: Generation and characterization of drug-target complex-specific antibodies
- Article: Antibodies for CAR-T Cell Therapy Development
- Poster: Faster Generation of Anti-Drug Antibodies Using SpyTag Technology
- Poster: CAR T cell analysis with modular antibodies
- Poster: Drug-Target-Complex Specific Antibodies for Pharmacokinetic Analysis of Biotherapeutics
- Poster: Characterization of anti-idiotypic antibodies for high performance in bioanalytical assays
- Poster: Recombinant anti-idiotypic antibodies for antibody drug development
- Poster: Generation of recombinant antibodies for Bio-Plex assays
-
Webinars, Videos and Technical Articles
-
Custom Recombinant Monoclonal Antibody Generation
s
Simplified sourcing via Scientist.com
s
Custom antibody project inquiry form
A personal, no obligation quotation for a custom monoclonal antibody generation project
s
Contact our custom antibody specialists
Tel: +49 (0) 89 80 90 95 45
Fax: +49 (0) 89 80 90 95 50
Office: Bio-Rad AbD Serotec GmbH, Campus Neuried, Anna-Sigmund-Str. 5, 82061 Neuried, Germany
Development of an assay to discriminate between native and truncated CXCL10 chemokine
Serial panning of the HuCAL PLATINUM® antibody library has isolated selective probes which permit differential quantification for two forms of the inflammatory chemokine CXCL10 in biological samples. The antibodies bind specifically to either native CXCL10 or an antagonist form, which is truncated by just two amino acids at the N-terminus. Assays with these antibodies have revealed high levels of antagonist chemokine in chronic HCV patients. Supplementary studies show dipeptidyl peptidase-4 (DPP4) is responsible for this cleavage, making it a putative target for adjuvant HCV therapeutics in non-responsive patients.
Recent investigations into the role of the chemokine CXCL10 (also known as IP-10) in hepatitis C virus (HCV) infection highlight the growing demand for highly selective antibodies in studies of inflammation and immunoregulation. While CXCL10 is known as a proinflammatory chemokine that is responsible for recruiting activated lymphocytes to the liver in HCV infected patients, elevated levels of CXCL10 were surprisingly shown to be a negative prognostic marker for response to therapy.
HCV infects nearly 170 million individuals worldwide. Studies show that the virus is cleared through HCV-specific CD8+ T cells, plus some NK cell activity. Curative treatment is successful in 40-80% of patients (numbers vary depending on viral genotype) with chronic infection following a combination regime of pegylated IFN-α2 and ribavirin.
The pathological mechanisms that prevent viral clearance are still uncertain, but the chemokine CXCL10 has been implicated. This circulating molecule binds to the cell surface receptor protein CXCR3 found especially on activated T lymphocytes and NK cells. A second step sees the flexible N-terminal tail of CXCL10 bind and trigger internal modulatory pathways. If active DPP4 cleaves the N-terminal tail, the CXCL10 molecule can still bind to the receptor but does not induce signaling. This form acts as an antagonist for the bioactive full length chemokine, thought to result in less powerful inflammatory responses.
A collaboration of French institutes solved this puzzle when they showed that blood and plasma samples from chronic HCV patients contained CXCL10 in an antagonistic form; this variant can still bind to its target receptor but does not trigger cell signaling. They also found that this antagonist form is a truncated version of active CXCL10, generated when the enzyme DPP4 (CD26) cleaves the first two amino acids from the N-terminus of the chemokine molecule. The scientists made their discoveries using recombinant antibodies isolated from the Human Combinatorial Antibody Library (HuCAL®). The specifically developed Fab antibodies showed high selectivity, binding either to full length CXCL10 or to the truncated chemokine, but not to both despite the close similarity between the two target forms. The team developed and validated ‘three-plex’ immunoassays, which they then used to investigate the in vitro and in vivo dynamics and activity of CXCL10 and DPP4.
About HuCAL
The HuCAL PLATINUM library contains more than 45 billion human antibodies against virtually every possible antigen or epitope. Sophisticated serial screening can isolate antibodies that have unique specificity for a native protein or cleavage product that may be truncated by just a few amino acids.
The HuCAL library has been created by mirroring the structural repertoire of the human immune system through the generation of master genes representing the various subclasses of human immunoglobulin. Sequences for the highly diversified complementarity determining regions (CDRs) of the antibody binding sites are synthesized by using trinucleotides. These ‘cassettes’ are combined to create antibody genes that code for some 45 billion unique fully human antibodies. HuCAL technology is well published and proven and has been used by the Bio-Rad custom antibody team to generate antibodies for research and diagnostic applications since 2004.
Selection of specific antibodies is performed in vitro, which allows researchers to generate single antibodies that can differentiate between antigens with minimal molecular differences such as the two amino acid truncation of CXCL10. As the sequence of any selected antibody is known, it is possible to reproduce the genes synthetically if needed. This sequence back up secures the future supply of the antibody and recombinant production methods ensure there is a high level of consistency between batches.
Guided selection strategies to identify antibodies against closely related antigens
Scientists from Bio-Rad isolated antibodies with selective binding to the agonist or antagonist forms of CXCL10. First, clones were isolated from the HuCAL library that recognized the truncated 7 amino acid N-terminal neoepitope but not the 9 amino acid peptide representing the native protein sequence. These antibodies were selected by three rounds of panning the library on the short peptide only (alternating between BSA and transferrin conjugates) and by blocking the library with the 9 amino acid peptide coupled to BSA (Figure 1).
To find the antibody specific for the full length protein and not the truncated form, an alternating panning was carried out on the 9 amino acid peptide coupled to BSA and the full length protein (Figure 2), to enrich antibodies binding to the folded protein. In this case the library was blocked with the 7 amino acid peptide coupled to BSA (not shown).
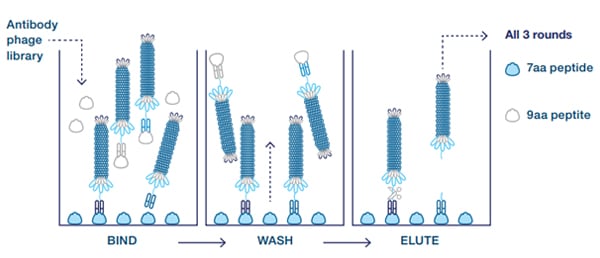
Fig. 1. Guided selection with blocking. The 9 amino acid peptide is used to remove crossreactive antibodies (counter-selection).
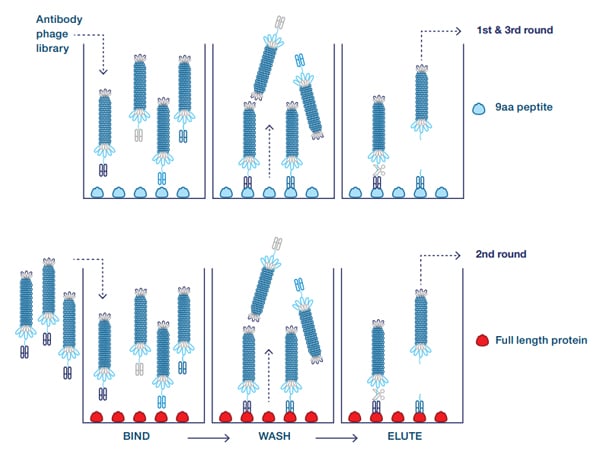
Fig. 2. Guided selection with alternating antigens. The antigen is alternated during the three rounds of selection, thereby enriching antibodies that bind to both antigens. Primary screening on both antigens is used to confirm the desired cross-reactivity.
Screening by direct ELISA was used to select the final clones for assay development, followed by sandwich ELISA to show antibody specificity to the whole and truncated versions of CXCL10 (Figure 3).
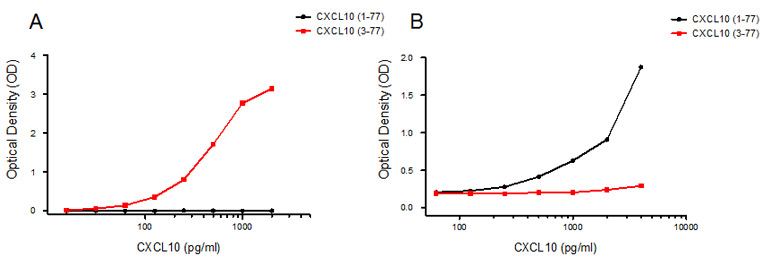
Fig. 3. Selection of antibody clones for assay development. A: Sandwich ELISA using capture antibody AbD09852, specific for the truncated version of CXCL10 (3-77). B: Sandwich ELISA using capture antibody AbD12010, specific for the long version of CXCL10 (1-77). Standard curves are shown using recombinant CXCL10 (1–77, black; 3–77, red).
The team used its isolated unique antibodies in fluorescent microsphere-based ‘three-plex’ assays that simultaneously quantified total, whole and truncated (antagonist) CXCL10. One of two assays developed by the team has been CLIA certified.
Key findings
The high specificity of the HuCAL-derived antibodies and their incorporation into a validated, quantified assay has enabled the team to publish the evidence that chemokine antagonism may contribute to some patients being unable to clear HCV even with treatment. Although CXCL10 secretion is high in these patients, elevated DPP4 activity leads to rapid cleavage of the N-terminal of CXCL10. This destroys the molecule’s ability to activate cell signaling pathways when it binds to its target immunoreceptor.
Interestingly, the differential assay also distinguished between the two forms of CXCL10 in urine samples taken from patients with non-muscle invasive bladder carcinoma being treated by intravesical bacilli Calmette-Guérin (BCG) therapy. Corresponding high levels of urinary DPP4 suggest that chemokine antagonism may also diminish the inflammatory response behind this treatment.
Conclusion
Biomarker discovery is high on the agenda for biomedical researchers as they search for more precise diagnostics and predictors for treatment outcomes. Validation of surrogate markers will help to improve clinical outcomes, support drug trials and provide an essential platform for more personalized medicine.
Post-translational modifications of a protein that result in different biologically relevant forms of the molecule are very important, especially when they impact activity or receptor binding. Antibodies that can distinguish between such closely related molecules are an essential tool for measurement of these changes. Isolation of recombinant antibodies using HuCAL technology provides a fast and effective way to generate such specialized reagents.
As demonstrated in this study, high quality antibodies isolated from the HuCAL library can discriminate between agonist and antagonist forms of CXCL10 in whole blood, plasma and urine samples. Evidence suggests that this chemokine is a strong predictor of poor clinical outcomes from HCV treatment. Deeper analysis points to DPP4 as a target for novel adjuvant therapeutics.
References
- Casrouge A et al. (2011). Evidence for an antagonist form of the chemokine CXCL10 in patients chronically infected with HCV. J Clin Invest 21, 308-317.
- Casrouge A et al. (2012). Discrimination of agonist and antagonist forms of CXCL10 in biological samples. Clin Exp Immunol 167, 137-148.
- Meissner et al. (2015). Dynamic Changes of Post-Translationally Modified Forms of CXCL10 and Soluble DPP4 in HCV Subjects Receiving Interferon-Free Therapy. Plos One DOI:10.1371/journal. pone.0133236.
Figure 3 is reproduced with permission from Casrouge A et al. (2012). Discrimination of agonist and antagonist forms of CXCL10 in biological samples. Clin Exp Immunol 167, 137-148. © 2011 The Authors. Clinical and Experimental Immunology © 2011 British Society for Immunology. Published by John Wiley and Sons.






